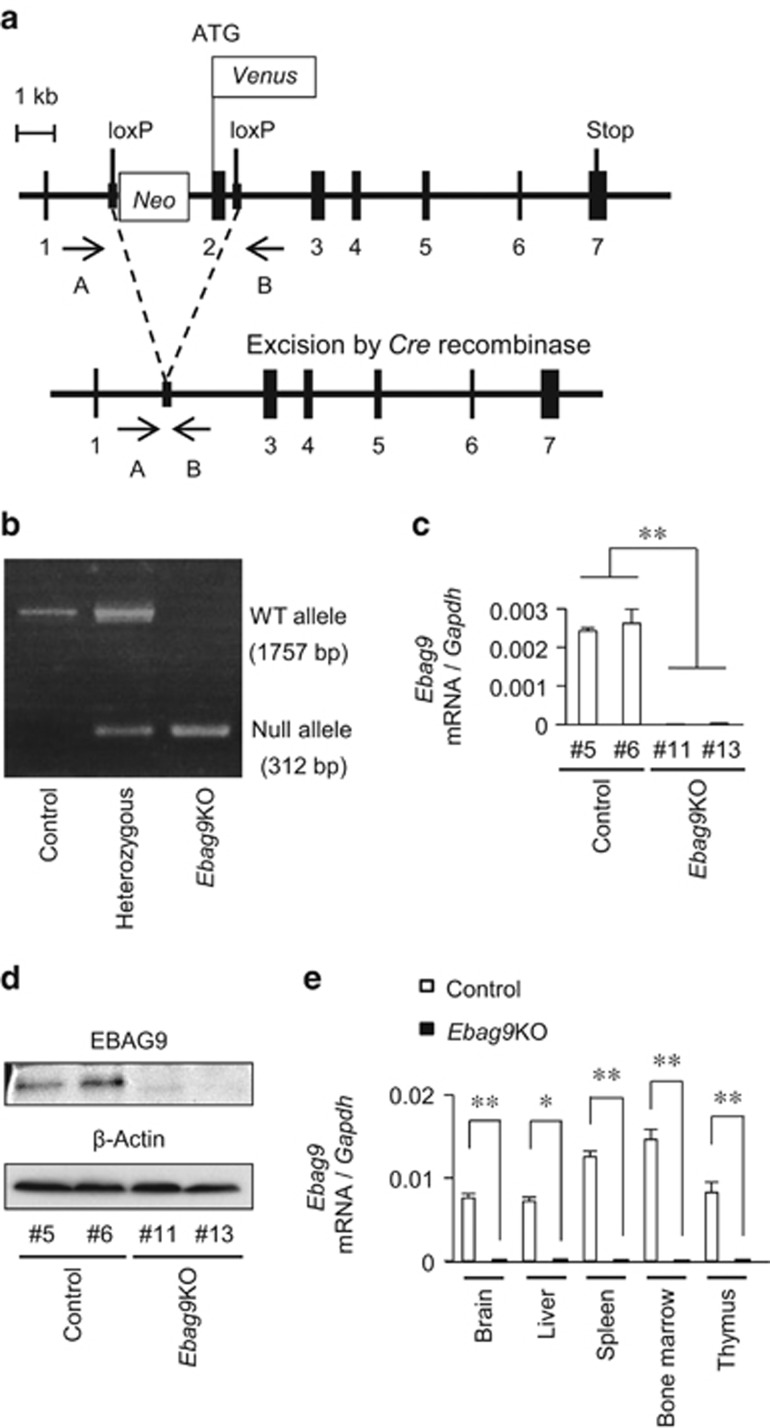
Figure 1.
Generation of Ebag9KO mice. (a) Schematic representation of the strategy employed to disrupt the Ebag9 gene. Ebag9f/f mice possess alleles including loxP sites flanking the neomycin-resistance cassette and Ebag9 exon 2 (top). Ebag9f/f mice were crossed with Ayu1-Cre mice. After removal of the loxP-flanked neomycin-resistance cassette via Cre-mediated recombination (bottom), the resulting offspring has a heterozygous deletion of Ebag9 (that is, Ebag9f/+;Ayu1-Cre). Ebag9f/+;Ayu1-Cre mice were bred to generate homozygous Ebag9f/f;Ayu1-Cre mice, designated as Ebag9KO mice. Positions of the PCR primers A and B, used for genotyping, are shown. (b) Representative PCR analysis of genomic DNA from mouse tails. Genotyping was performed by PCR using primers A and B for the wild-type allele and the null allele. The genotypes and their corresponding PCR products are as follows: control (Ebag9+/+;Ayu1-Cre), 1757 bp; heterozygous (Ebag9f/+;Ayu1-Cre), 1757 and 312 bp; Ebag9KO (Ebag9f/f;Ayu1-Cre), 312 bp. (c) qRT–PCR analysis of full-length Ebag9 mRNA expression in MEFs obtained from Ebag9KO and control mice. Gapdh was used as an internal control. (d) Western blot analysis of EBAG9 protein expression in MEFs obtained from Ebag9KO and control mice. β-Actin was used as a loading control. (e) Ebag9 mRNA expression in several tissues derived from Ebag9KO and control mice. Ebag9 mRNA expression was analyzed by qRT–PCR. Gapdh was used as an internal control. The results shown are mean values±s.d. *P<0.05; **P<0.01 by Student's t-test.