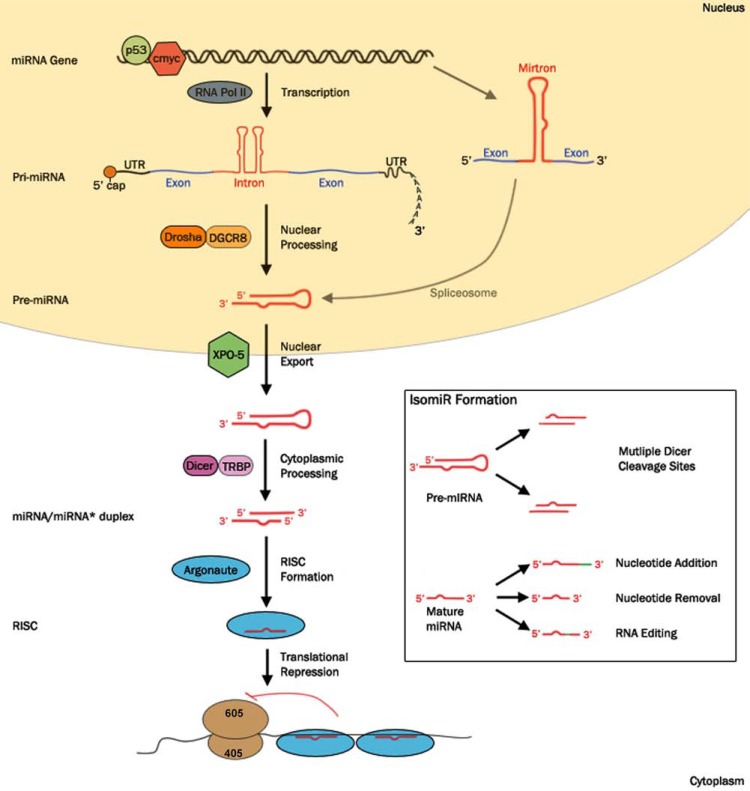
Fig. (1).
miRNA biosynthesis and isomiR formation. Canonical maturation of mammalian miRNAs begins with the miRNA-containing gene being transcribed to form the primary transcript (pri-miRNA). This transcription, a function of RNA polymerase II (RNA Pol II), can be regulated by transcription factors implicated in cancer pathogenesis e.g. p53, c-myc. Pri-miRNA is cleaved by the microprocessor complex (Drosha-DGCR8) to form the precursor miRNA (pre-miRNA) which is then exported into the cytoplasm by Exportin-5 (XPO-5). The protein complex Dicer-TRBP cleaves the pre-miRNA to form the miRNA/miRNA* duplex. One strand of this duplex, the guide strand, is then loaded into an Argonaute protein to form the RNA-Induced Silencing Complex (RISC). The mature miRNA guides the RISC to mRNA targets, resulting in the translation repression of the bound transcripts. Alternative miRNA biosynthesis pathways includes the production of mirtrons, miRNAs that derive from short intronic hairpins, produced from precursors formed by the splicing machinery.
Inset. IsomiRs describe miRNA variants that are produced from a common miRNA gene. Typical sources of isomiRs include multiple Dicer cleavage sites present within the pre-miRNA hairpin. Additionally, isomiRs are formed via post-transcriptional modifications of the mature miRNA, such as the addition or removal of nucleotides, or changing internal nucleotides via RNA editing.