Significance
We present highlights of the first complete domestic cat reference genome, to our knowledge. We provide evolutionary assessments of the feline protein-coding genome, population genetic discoveries surrounding domestication, and a resource of domestic cat genetic variants. These analyses span broadly, from carnivore adaptations for hunting behavior to comparative odorant and chemical detection abilities between cats and dogs. We describe how segregating genetic variation in pigmentation phenotypes has reached fixation within a single breed, and also highlight the genomic differences between domestic cats and wildcats. Specifically, the signatures of selection in the domestic cat genome are linked to genes associated with gene knockout models affecting memory, fear-conditioning behavior, and stimulus-reward learning, and potentially point to the processes by which cats became domesticated.
Keywords: Felis catus, domestication, genome
Abstract
Little is known about the genetic changes that distinguish domestic cat populations from their wild progenitors. Here we describe a high-quality domestic cat reference genome assembly and comparative inferences made with other cat breeds, wildcats, and other mammals. Based upon these comparisons, we identified positively selected genes enriched for genes involved in lipid metabolism that underpin adaptations to a hypercarnivorous diet. We also found positive selection signals within genes underlying sensory processes, especially those affecting vision and hearing in the carnivore lineage. We observed an evolutionary tradeoff between functional olfactory and vomeronasal receptor gene repertoires in the cat and dog genomes, with an expansion of the feline chemosensory system for detecting pheromones at the expense of odorant detection. Genomic regions harboring signatures of natural selection that distinguish domestic cats from their wild congeners are enriched in neural crest-related genes associated with behavior and reward in mouse models, as predicted by the domestication syndrome hypothesis. Our description of a previously unidentified allele for the gloving pigmentation pattern found in the Birman breed supports the hypothesis that cat breeds experienced strong selection on specific mutations drawn from random bred populations. Collectively, these findings provide insight into how the process of domestication altered the ancestral wildcat genome and build a resource for future disease mapping and phylogenomic studies across all members of the Felidae.
The domestic cat (Felis silvestris catus) is a popular pet species, with as many as 600 million individuals worldwide (1). Cats and other members of Carnivora last shared a common ancestor with humans ∼92 million years ago (2, 3). The cat family Felidae includes ∼38 species that are widely distributed across the world, inhabiting diverse ecological niches that have resulted in divergent morphological and behavioral adaptations (4). The earliest archaeological evidence for human coexistence with cats dates to ∼9.5 kya in Cyprus and ∼5 kya in central China (5, 6), during periods when human populations adopted more agricultural lifestyles. Given their sustained beneficial role surrounding vermin control since the human transition to agriculture, any selective forces acting on cats may have been minimal subsequent to their domestication. Unlike many other domesticated mammals bred for food, herding, hunting, or security, most of the 30–40 cat breeds originated recently, within the past 150 y, largely due to selection for aesthetic rather than functional traits.
Previous studies have assessed breed differentiation (6, 7), phylogenetic origins of the domestic cat (8), and the extent of recent introgression between domestic cats and wildcats (9, 10). However, little is known regarding the impact of the domestication process within the genomes of modern cats and how this compares with genetic changes accompanying selection identified in other domesticated companion animal species. Here we describe, to our knowledge, the first high-quality annotation of the complete domestic cat genome and a comparative genomic analysis including whole-genome sequences from other felids and mammals to identify the molecular footprints of the domestication process within cats.
Results and Discussion
To identify molecular signatures underlying felid phenotypic innovations, we developed a higher-quality reference assembly for the domestic cat genome using whole-genome shotgun sequences (Materials and Methods and SI Materials and Methods). The assembly (FelCat5) comprises 2.35 gigabases (Gb) assigned to all 18 autosomes and the X chromosome relying on physical and linkage maps (11) with a further 11 megabases (Mb) in unplaced scaffolds. The assembly is represented by an N50 contig length of 20.6 kb and a scaffold N50 of 4.7 Mb, both of which show substantial improvement over previous light-coverage genome survey sequences that included only 60% of the genome (12, 13). The Felis catus genome is predicted to contain 19,493 protein-coding genes and 1,855 noncoding RNAs, similar to dog (14). Hundreds of feline traits and disease pathologies (15) offer novel opportunities to explore the genetic basis of simple and complex traits, host susceptibility to infectious diseases, as well as the distinctive genetic changes accompanying the evolution of carnivorans from other mammals.
To identify signatures of natural selection along the lineages leading to the domestic cat, we identified rates of evolution using genome-wide analyses of the ratio of divergence at nonsynonymous and synonymous sites (dN/dS) (16) (Materials and Methods and SI Materials and Methods). We used the annotated gene set (19,493 protein-coding genes) to compare unambiguous mammalian gene orthologs shared between cat, tiger, dog, cow, and human (n = 10,317). Two-branch and branch-site models (17) collectively identified 467, 331, and 281 genes that were putatively shaped by positive selection in the carnivore, felid, and domestic cat (subfamily Felinae) ancestral lineages, respectively (S1.1–S1.3 in Dataset S1). We assessed the potential impact of amino acid changes using TreeSAAP (18) and PROVEAN (19). The majority of identified genes possess substitutions with significant predicted structural or biochemical effects based on one or both tests (Fig. S1 and S1.4 in Dataset S1). Although the inferences produced by our methods call for additional functional analyses, we highlight several positively selected genes to illustrate their importance to carnivore and feline biology.
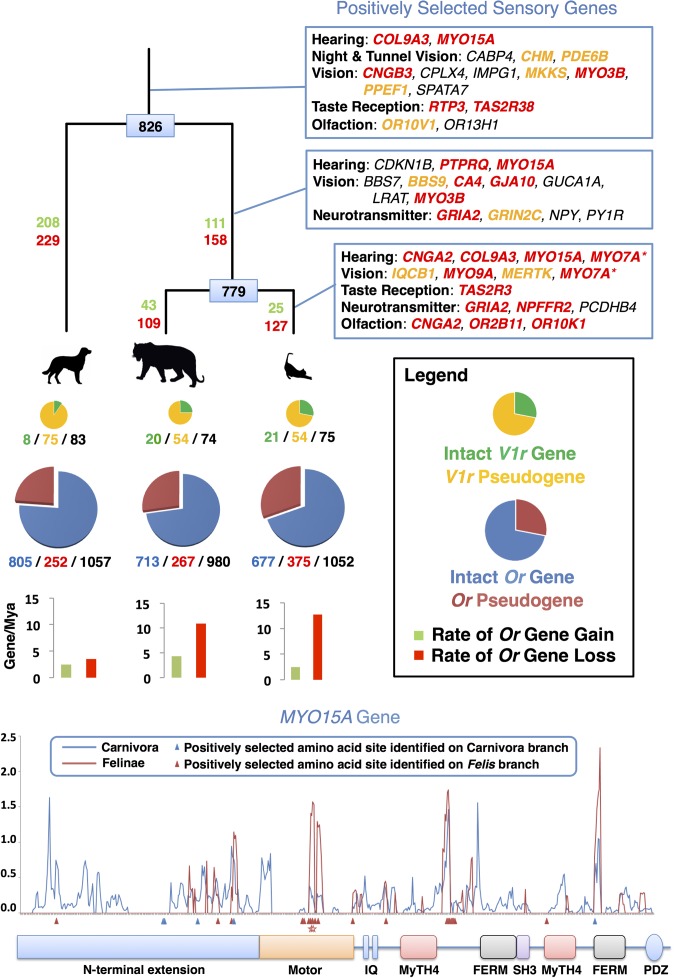
Carnivores are endowed with extremely acute sensory adaptations, allowing them to effectively locate potential prey before being discovered (20). Within carnivores, cats have the broadest hearing range, allowing them to detect both ultrasonic communication by prey as well as their movement (21). We identified six positively selected genes (Fig. 1) that conceivably evolved to increase auditory acuity over a wider range of frequencies in the carnivore ancestor and within Felidae, as mutations within each gene have been associated with autosomal, nonsyndromic deafness or hearing loss (22, 23). Visual acuity is adaptive for hunting and catching prey, especially for crepuscular predators such as the cat and other carnivores. Accordingly, we identified elevated dN/dS values for 20 carnivoran genes that, when mutated in humans, have well-described roles in a spectrum of visual pathologies (Fig. 1). For example, truncating mutations in human CHM cause the progressive disease choroideremia (24), beginning with a loss of night vision and peripheral vision and later a loss of central vision. Many carnivores have excellent night vision (20, 25), and we postulate that the acquisition of selectively advantageous amino acid substitutions within several genes increased visual acuity under low-light conditions. In one interesting dual-role example, MYO7A encodes a protein involved in the maintenance of both auditory and visual systems that, when mutated, results in loss of hearing and vision (26).
Fig. 1.
Dynamic evolution of feline sensory repertoires (Upper). The phylogenetic tree depicts relationships scaled to time between dog, tiger, and domestic cat. Positively selected genes are listed (Top Right), with lines indicating genes identified on the ancestral branch of Carnivora (Top), Felidae (Middle), and Felinae (Bottom). Genes highlighted in red and orange were identified with significant structural or biochemical effects by two tests or one test, respectively (S1.4 in Dataset S1). MYO7A (*) expression is associated with hearing and vision. Numbers at each tree node represent the reconstructed ancestral functional olfactory receptor gene (Or) repertoire for carnivores and felids. Numbers labeling each branch are estimated Or gene gain (green) and loss (red). The pie charts refer to functional and nonfunctional (pseudogenic) vomeronasal (V1r; Top) and Or (Bottom) gene repertoires, with circles drawn in proportion to the size of each gene repertoire. Or genes are depicted in blue (functional) and red (nonfunctional), and V1r genes are depicted in green (functional) and yellow (nonfunctional). Beneath each pie chart are numbers of functional/nonfunctional/total genes identified in the current genome annotations of the three species. Bar graphs depict rates of Or gene gain and loss. Location of signatures of positive selection (Lower). Several genes encode members of the myosin gene family of mechanochemical proteins, with MYO15A notably under selection in all three branches tested. Curved lines represent the estimated dN/dS values (y axis) calculated in 90-bp sliding windows (step size of 18 bp) along the length of the gene alignment (x axis) for dog, cat, and tiger. Colored boxes indicate known functional domains. Arrowheads indicate the location of positively selected amino acid sites based on the results of the branch-site test. Stars indicate deleterious mutations in the domestic cat (Materials and Methods). Motifs and domains include the IQ calmodulin-binding motif (IQ); the myosin tail homology 4 domain (MyTH4); the FERM domain (FERM); the SRC homology 3 domain (SH3); and the PDZ domain (PDZ).
Cats differ from most other carnivores as a result of being obligately carnivorous. One outcome of this adaptive process is that cats are unable to synthesize certain essential fatty acids, specifically arachidonic acid, due to low Delta-6-desaturase activity (27). This has led to suggestions that cats use an alternate (yet unknown) pathway to generate this essential fatty acid for normal health and reproduction. Furthermore, cats fed a diet rich in saturated and polyunsaturated fatty acids showed no effects on plasma lipid concentrations that in humans are risk factors for coronary heart disease and atherosclerosis (28). These aspects of feline biology are reflected in our positive selection results, where the notable classes of genes overrepresented in the Felinae list are related to lipid metabolism (S1.5 in Dataset S1). For example, one of the positively selected genes, ACOX2, is critical for metabolism of branch-chain fatty acids and has been suggested to regulate triglyceride levels (29), whereas mutations in PAFAH2 have been associated with risk for coronary heart disease and ischemia (30). The enrichment of genes related to lipid metabolism is likely a signature of adaptation for accommodating the hypercarnivorous diet of felids (31), and mirrors similar signs of selection on lipid metabolic pathways in the genomes of polar bears (32).
Gene duplication and gene loss events often play substantial roles in phenotypic differences between species. To identify protein families that rapidly evolved in the domestic cat, either by contraction or expansion, we examined gene family expansion along an established species tree (33) using tree orthology (34). Two extensive chemosensory gene families, coding for olfactory (Or) and vomeronasal (V1r) receptors, are responsible for small-molecule detection of odorants and other chemicals for mediating pheromone perception, respectively. Cats rely less on smell to hunt and locate prey in comparison with dogs, which are well-known for their olfactory prowess (35). These observations are confirmed by our analysis of the complete Or gene repertoires for cat, tiger, and dog (Fig. 1), illustrating smaller functional repertoires in felids relative to dogs (∼700 genes versus >800, respectively). By contrast, the V1r gene repertoire is markedly reduced in dogs but expanded in the ancestor of the cat family (8 versus 21 functional genes, respectively), with evidence for species-specific gene loss in different felids (Fig. 1 and Figs. S2 and S3). A growing body of evidence cataloging Or gene repertoires in diverse mammals demonstrates common tradeoffs between functional Or repertoire size and other sensory systems involved in ecological niche specialization, such as loss of Or genes coinciding with gains in trichromatic color vision in primates (36) and chemosensation in platypus (37). These results add further evidence supporting cats’ extensive reliance on pheromones for sociochemical communication (38), which is consistent with a genomic tradeoff between functional Or and V1r repertoires in response to uniquely evolved ecological strategies in the canid and felid lineages (4).
Cats are considered only a semidomesticated species, because many populations are not isolated from wildcats and humans do not control their food supply or breeding (39, 40). We therefore predicted a relatively modest effect of domestication on the cat genome based on recent divergence from and ongoing admixture with wildcats (8–10), a relatively short human cohabitation time compared with dogs (5, 6), and the lack of clear morphological and behavioral differences from wildcats, with docility, gracility, and pigmentation being the exceptions. To identify genomic regions showing signatures of selection influenced by the domestication process, we used whole-genome analyses of cats from different domestic breeds and wildcats (i.e., other F. silvestris subspecies) using pooling methods that control for genetic drift (41). Detecting the genomic regions under putative selection during cat domestication can be complicated by random fixation due to genetic drift during the formation of breeds. We mitigated this effect by combining sequence data from a collection of 22 cats (∼58× coverage) from six phylogenetically and geographically dispersed domestic breeds (42) before variant detection and performed selection analyses relative to variants detected within a pool of European (F. silvestris silvestris) and Near Eastern (F. silvestris lybica) wildcats (∼7× coverage; Figs. S4 and S5 and S2.1 in Dataset S2).
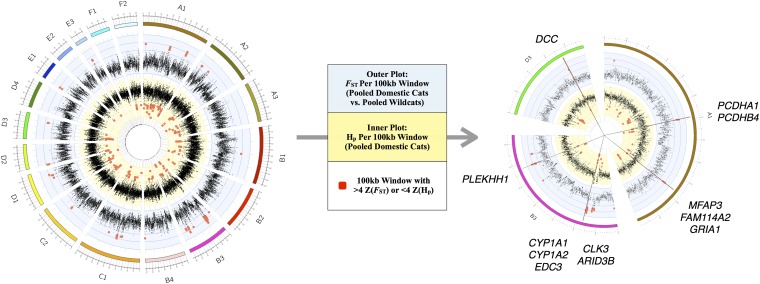
After stringent filtering of resequencing data, we aligned sequences to the cat reference genome and identified 8,676,486 and 5,190,430 high-quality single-nucleotide variants (SNVs) among domestic breeds and wildcats, respectively, at a total of 10,975,197 sites (Fig. S3). We next identified 130 regions along cat autosomes with either pooled heterozygosity (Hp) 4 SDs below the mean or divergence (FST) greater than 4 SDs from the mean (Figs. S4 and S6, SI Materials and Methods, and S2.2 and S2.3 in Dataset S2). After parsing regions of high confidence displaying both low domestic Hp and high FST, we found 13 genes underlying five chromosomal regions (Fig. 2, Fig. S4, and S2.4 in Dataset S2). Genes within each of these regions play important roles in neural processes, notably pathways related to synaptic circuitry that influence behavior and contextual clues related to reward.
Fig. 2.
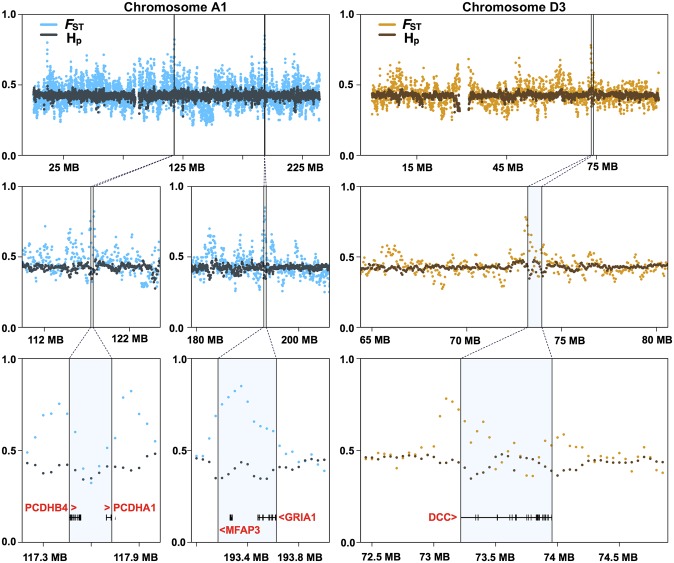
Sliding window analyses identify five regions of putative selection in the domestic cat genome. Measurements of Z-transformed pooled heterozygosity in cat [inner plot; Z(Hp)] and the Z-transformed fixation index between pooled domestic cat and pooled wildcat [outer plot; Z(FST)] for autosomal 100-kb windows across all 18 autosomes (Left). Red points indicate windows that passed the threshold for elevated divergence [>4 Z(FST)] or low diversity [<4 Z(HP)]. The five regions of putative selection are represented by the straight lines and include contiguous windows that passed both thresholds for elevated divergence and low diversity (Right). These regions, across cat autosomes A1, B3, and D3, contain 12 known genes.
One putative region of selection along chromosome A1 (chrA1) (Fig. 3) is denoted by a pair of protocadherin genes (PCDHA1 and PCDHB4), which establish and maintain specific neuronal connections and have implications for synaptic specificity, serotonergic innervation of the brain, and fear conditioning (43). PCDHB4 was also identified in the dN/dS analyses. A second region, also on chrA1 (Fig. 3), overlaps with a glutamate receptor gene, GRIA1. Glutamate receptors are the predominant excitatory neurotransmitter receptors in the mammalian brain and play an important role in the expression of long-term potentiation and memory formation (44). GRIA1 knockout mice exhibit defects in stimulus-reward learning, notably those related to food rewards (45). Two additional glutamate receptor genes, GRIA2 and NPFFR2, have elevated dN/dS rates within the domestic cat branch of the felid tree (Fig. 1). A third region on chromosome D3 (Fig. 3) encompasses a single gene, DCC, encoding the netrin receptor. This gene shows abundant expression in dopaminergic neurons, and behavioral studies of DCC-deficient mice show altered dopaminergic system organization, culminating in impaired memory, behavior, and reward responses (46, 47). Two additional regions on chromosome B3 harbor strong signatures of selection (Fig. S7). The first contains three genes, including ARID3B (AT rich interactive domain 3B), which plays a critical role in neural crest cell survival (48). The second region contains a single gene, PLEKHH1, which encodes a plekstrin homology domain expressed predominantly in human brain. Human genome-wide association studies link variants in PLEKHH1 with sphingolipid concentrations that, when altered, lead to neurological and psychiatric disease (49).
Fig. 3.
Comparison between domestic cats and wildcats identifying genes within putative regions of selection in the domestic cat genome that are associated with pathways related to synaptic circuitry and contextual clues related to reward. We identified 130 regions along cat autosomes with either pooled domestic Z(Hp) < −4 or Z(FST) > 4, and 5 annotated regions met both criteria. A total of 12 genes was found within these regions, many of which are implicated in neural processes; for instance, genes within regions along chromosomes A1 and D3 are highlighted.
The genetic signals from this analysis fall in line with the predictions of the domestication syndrome hypothesis (50), which posits that the morphological and physiological traits modified by mammalian domestication are explained by direct and indirect consequences of mild neural crest cell deficits during embryonic development. ARID3B, DCC, PLEKHH1, and protocadherins are all implicated in neural crest cell migration. ARID3B is induced in developing mouse embryos during the differentiation of neural crest cells to mature sympathetic ganglia cells (51). DCC directly interacts with the Myosin Tail Homology 4 (MyTH4) domain of MYO10 (myosin X) (52), a gene critical for the migratory ability of neural crest cells. In this way, DCC regulates the function of MYO10 to stimulate the formation and elongation of axons and cranial neural crest cells in developing mouse (53) and frog embryos (54). Like MYO10, PLEKHH1 contains a MyTH4 domain and interacts with the transcription factor MYC, a regulator of neural crest cells, to activate transcription of growth-related genes (55). Taken together, we propose that changes in these neural crest-related genes underlie the evolution of tameness during cat domestication, in agreement with analyses of other domesticated genomes (56–58).
We also examined regions of high genetic differentiation between domestic cats and wildcats and observed enrichment in several Wiki and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (S2.5 in Dataset S2), including homologous recombination and axon guidance. Divergence in regions harboring homologous recombination genes (RAD51B, ZFYVE26, BRCA2) may contribute to the high recombination rate reported for domestic cats relative to other mammals (59). Previous studies have suggested that domestication may select for an increase in recombination as a mechanism to generate diversity (60). Specifically, selection for a recombination driver allele may be favored when it is tightly linked to two or more genes with alleles under selection (61). We hypothesize that the close proximity (<350 kb) of two adjacent genes that regulate homologous recombination (ZFYVE26 and RAD51B, which directly interact with BRCA2), two visual genes (RDH11 and RDH12) related to retinol metabolism and dark adaptation (62), and one of our candidate domestication genes, PLEKHH1 (S2.4 in Dataset S2), represents such a case of adaptive linkage.
Aesthetic qualities such as hair color, texture, and pattern strongly differentiate wildcats from domesticated populations and breeds; however, unlike other domesticated species, less than 30–40 genetically distinct breeds exist (63). At the beginning of the cat fancy ∼200 y ago, only five different cat “breeds” were recognized, with each being akin to geographical isolates (64). Long hair and the Siamese coloration of “points” were the only diagnostic breed characteristics. Although most breeds were developed recently, following different breeding strategies and selection pressures, much of the color variation in cats developed during domestication, before breed development, and thus is known as “natural” or “ancient” mutations by cat fanciers.
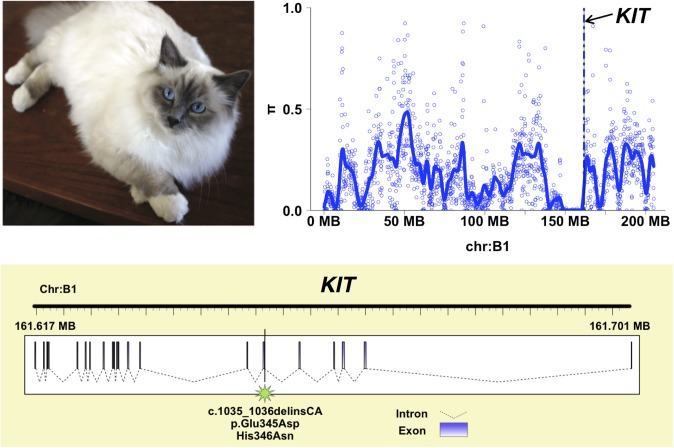
White-spotting phenotypes are a hallmark of domestication, and in cats can range from a complete lack of pigmentation (white) to intermediate bicolor spotting phenotypes (spotting) to white at only the extremities (gloving). For instance, the Birman breed is characterized by point coloration, long hair, and gloving (Fig. 4). A recent study in several white-spotted cats localized the mutation responsible for the spotting pigmentation phenotype within KIT intron 1 (65). The KIT gene, located on cat chromosome B1 (66), is primarily involved in melanocyte migration, proliferation, and survival (67). Surprisingly, direct PCR and sequencing excluded the published dominant allele as being associated with the white coloration pattern in Birman (SI Materials and Methods). At the same time, whole-genome resequencing data from a pooled sample of Birman cats (n = 4; SI Materials and Methods and S2.6 in Dataset S2) identified the genomic region containing KIT as an outlier exhibiting unusually low genetic diversity (Fig. 4). We therefore resequenced KIT exons in a large cohort of domestic cats with various white-spotting phenotypes to genotype candidate SNVs (409 from 21 breeds, 5 Birman outcrosses, and 315 random bred cats). We identified just two adjacent missense mutations that were concordant with the gloving pattern in Birman cats (Fig. 4 and S2.7 in Dataset S2). Genotyping these SNPs in a larger sample including 150 Birman cats and 729 additional cats confirmed that all Birman cats were homozygous for both SNPs and that all first-generation outcrossed Birman cats with no gloving were carriers of the polymorphisms (S2.8 in Dataset S2).
Fig. 4.
Genetics of the gloving pigmentation pattern in the Birman cat. The paws of the Birman breed (Top Left) are distinguished by white gloving. The average nucleotide diversity adjacent to KIT was low (Top Right). Sequencing experiments identified two adjacent missense mutations within exon 6 of KIT that were concordant with the gloving pattern in Birman cats (Bottom).
Several lines of evidence indicate that the gloving phenotype in the Birman breed is the result of these two recessive mutations in KIT. Both mutations affect the fourth Ig domain of KIT, and mutations in this motif near the dimerization site have been shown to result in accelerated ligand dissociation and reduced downstream signal transduction events (68). Interestingly, the frequency of the Birman gloving haplotype in the Ragdoll breed, which shares an extremely similar white-spotting phenotype, was only 12.3%. We suggest that other genetic variants, including the endogenous retrovirus insertion in KIT intron 1 (65), likely contribute to the white-spotting phenotype in the Ragdoll breed. The frequency of the Birman gloving haplotype is just 10% in the random nonbreed population, thus illustrating a case where segregating genetic variation in ancestral nonbred populations has reached fixation within Birman cats through strong artificial selection in a remarkably short time frame.
In conclusion, our analyses have identified genetic signatures within feline genomes that match their unique biology and sensory skills. The number of genomic regions with strong signals of selection since cat domestication appears modest compared with those in the domestic dog (41), which is concordant with a more recent domestication history, the absence of strong selection for specific physical characteristics, as well as limited isolation from wild populations. Our results suggest that selection for docility, as a result of becoming accustomed to humans for food rewards, was most likely the major force that altered the first domesticated cat genomes.
Materials and Methods
A female Abyssinian cat, named Cinnamon, served as the DNA source for all sequencing reads (12). From this source we generated ∼14× whole-genome shotgun coverage with Sanger and 454 technology. A BAC library was also constructed and all BACs were end-sequenced. We assembled the combined sequences using CABOG software (69) (SI Materials and Methods).
We estimated nonsynonymous and synonymous substitution rates using the software PAML 4.0 (17). The following pipeline was used to perform genome-wide selection analyses. (i) We identified 10,317 sets of 1:1:1:1:1 orthologs from the whole-genome annotations of human (GRCh37), cow (UMD3.1), dog (CanFam3.1), tiger (tigergenome.org), and domestic cat using the Ensembl pipeline (70). We tested for signatures of natural selection assuming the species tree topology (((cat, tiger), dog), cow, human). (ii) We aligned the translated amino acid sequence of the coding region of each gene using MAFFT (71) with the slow and most accurate parameter settings. A locally developed Perl script pipeline was applied that removed poorly aligned or incorrectly annotated amino acid residues caused by obvious gene annotation errors within the domestic cat and tiger genome assemblies. Aligned amino acid sequences were used for guiding nucleotide-coding sequences by adding insertion gaps and removing poorly aligned regions. (iii) Model testing and likelihood ratio tests (LRTs) were performed using PAML 4.0. Paired models representing different hypotheses consisted of branch tests and branch-site tests (fixed ω = 1 vs. variable ω). For the branch-specific tests, free ratio vs. one-ratio tests were used to identify putatively positively selected genes. These genes were subsequently tested by two-ratio and one-ratio models to identify genes with significant positive selection of one branch versus all other branches (two-branch test). Significance of LRT results used a threshold of P < 0.05. We also report the mean synonymous rates along the ancestral felid lineage as well as the tiger, cat, and dog lineages (Fig. S1). We assessed enrichment of gene functional clusters under positive natural selection using WebGestalt (72) (S1.5–S1.7 in Dataset S1). Entrez Gene IDs were input as gene symbols, with the organism of interest set to Homo sapiens using the genome as the reference set. Significant Gene Ontology categories (73), Pathway Commons categories (74), WikiPathways (75), and KEGG Pathways (76) were reported using a hypergeometric test, and the significance level was set at 0.05. We implemented the Benjamini and Hochberg multiple test adjustment (77) to control for false discovery.
Using the whole-genome assembly of domestic cat (FelCat5) as a reference, we mapped Illumina raw sequences from a pool of four wildcat individuals [two European wildcats (F. s. silvestris) and two Eastern wildcats (F. s. lybica)]. Six additional domestic cat breeds from different worldwide regional populations were sequenced using the Illumina platform (SI Materials and Methods). Before sequencing, we pooled samples by breed for the following individuals: Maine Coon (n = 5), Norwegian Forest (n = 4), Birman (n = 4), Japanese Bobtail (n = 4), and Turkish Van (n = 4). Whole-genome sequencing was also performed on an Egyptian Mau cat (n = 1) and on the Abyssinian reference individual (n = 1).
We combined the raw reads from the following breed sequencing experiments (described above) before alignment and variant calling: Egyptian Mau, Maine Coon, Norwegian Forest, Birman, Japanese Bobtail, and Turkish Van. The domestic cat pool (n = 22) was sequenced to a genome coverage depth of ∼58-fold, whereas the wildcat pool was sequenced to a depth of ∼7-fold (S2.1 in Dataset S2). Base position differences were called using the convergent outcomes of the software SAMtools (78) and VarScan 2 (79). Parameters included a P value of 0.1, a map quality of 10, and parameters for filtering by false positives. A clustered variant filter was implemented to allow for a maximum of five variant sites in any 500-bp window. Variants were finally filtered using PoPoolation2 (80) to yield a high-confidence set of SNVs (n = 6,534,957; filtering steps included a minimum coverage of 8, a minimum variant count of 6, a maximum coverage of 500 for the domestic cat pool, and a maximum coverage of 200 for the wildcat pool).
We screened for positively selected candidate genes during cat domestication by parsing specific 100-kb windows that showed low diversity [low pooled heterozygosity (Hp)] in domestic cat breeds and had high divergence [a high fixation index (FST)] between domestic cats and wildcats (41, 81). FST was calculated using PoPoolation2, and measurements of Hp were calculated using a custom script. A total of 6,534,957 high-quality SNV sites were used to calculate FST and Hp at each 100-kb window, and a step size of 50 kb was incorporated. All windows containing less than 10 variant sites were removed from the analysis, resulting in n = 46,906 100-kb windows along cat autosomes, as represented in the FelCat5 assembly. We Z-transformed the autosomal Hp [Z(Hp)] and FST [Z(FST)] distributions and designated as putatively selected regions those that fell at least 4 SDs away from the mean [Z(Hp) < −4 and Z(FST) > 4]. We applied a threshold of Z(Hp) ≤ −4 and Z(FST) ≥ 4 for putative selective sweeps, because windows below or above these thresholds represent the extreme lower and extreme upper ends of the respective distributions (Fig. S4). Windows with elevated FST or depressed Hp were annotated for gene content using the intersect tool in BEDTools (82). Enrichment analysis of underlying gene content was carried out using WebGestalt (72) using the same methods as described above, except only significant WikiPathways (75) and KEGG Pathways (76) were reported (S2.5 and S2.10–S2.11 in Dataset S2).
Primers to amplify KIT exons (ENSFCAG00000003112) were designed using Primer3Plus (83) and annealed to intronic regions flanking each exon. A PCR assay was performed to determine the presence or absence of the dominant, white-spotting retroviral insertion in KIT (65). An allele-specific PCR assay was designed for genotyping exon 6 SNPs (S2.9 in Dataset S2). See SI Materials and Methods for additional details.
Supplementary Material
Acknowledgments
We thank The Genome Institute members Kim Kyung, Dave Larson, Karyn Meltz Steinburg, and Chad Tomlinson for providing assistance and advice on data analysis, and Tom Nicholas for manuscript review. We also thank NIH/National Institute on Alcohol Abuse and Alcoholism members David Goldman and Qiaoping Yuan. The cat genome and transcriptome sequencing was funded by NIH/National Human Genome Research Institute Grant U54HG003079 (to R.K.W.). Further research support included grants to M.W.H. (National Science Foundation Grant DBI-0845494), W.J.M. (Morris Animal Foundation Grants D06FE-063 and D12FE-019), T.M.-B. (European Research Council Starting Grant 260372 and Spanish Government Grant BFU2011-28549), and L.A.L. [National Center for Research Resources (R24 RR016094), Office of Research Infrastructure Programs/Office of the Director (R24 OD010928), and Winn Feline Foundation (W10-014, W09-009)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GU270865.1, KJ923925–KJ924979, SRX026946, SRX026943, SRX026929, SRX027004, SRX026944, SRX026941, SRX026909, SRX026901, SRX026955, SRX026947, SRX026911, SRX026910, SRX026948, SRX026928, SRX026912, SRX026942, SRX026930, SRX026913, SRX019549, SRX019524, SRX026956, SRX026945, and SRX026960).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410083111/-/DCSupplemental.
References
- 1.American Pet Product Manufacturing Association . National Pet Owner’s Survey. Am Pet Prod Manuf Assoc; Greenwich, CT: 2008. [Google Scholar]
- 2.Meredith RW, et al. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science. 2011;334(6055):521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 3.Hedges SB, Dudley J, Kumar S. TimeTree: A public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22(23):2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 4.Sunquist M, Sunquist F. Wild Cats of the World. Univ of Chicago Press; Chicago: 2002. [Google Scholar]
- 5.Vigne J-D, Guilaine J, Debue K, Haye L, Gérard P. Early taming of the cat in Cyprus. Science. 2004;304(5668):259. doi: 10.1126/science.1095335. [DOI] [PubMed] [Google Scholar]
- 6.Hu Y, et al. Earliest evidence for commensal processes of cat domestication. Proc Natl Acad Sci USA. 2014;111(1):116–120. doi: 10.1073/pnas.1311439110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menotti-Raymond M, et al. Patterns of molecular genetic variation among cat breeds. Genomics. 2008;91(1):1–11. doi: 10.1016/j.ygeno.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Driscoll CA, et al. The Near Eastern origin of cat domestication. Science. 2007;317(5837):519–523. doi: 10.1126/science.1139518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nussberger B, Greminger MP, Grossen C, Keller LF, Wandeler P. Development of SNP markers identifying European wildcats, domestic cats, and their admixed progeny. Mol Ecol Resour. 2013;13(3):447–460. doi: 10.1111/1755-0998.12075. [DOI] [PubMed] [Google Scholar]
- 10.Beaumont M, et al. Genetic diversity and introgression in the Scottish wildcat. Mol Ecol. 2001;10(2):319–336. doi: 10.1046/j.1365-294x.2001.01196.x. [DOI] [PubMed] [Google Scholar]
- 11.Bach LH, et al. A high-resolution 15,000Rad radiation hybrid panel for the domestic cat. Cytogenet Genome Res. 2012;137(1):7–14. doi: 10.1159/000339416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pontius JU, et al. Agencourt Sequencing Team NISC Comparative Sequencing Program Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17(11):1675–1689. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mullikin JC, et al. NISC Comparative Sequencing Program Light whole genome sequence for SNP discovery across domestic cat breeds. BMC Genomics. 2010;11(1):406. doi: 10.1186/1471-2164-11-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438(7069):803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 15.Nicholas FW. Online Mendelian Inheritance in Animals (OMIA): A comparative knowledgebase of genetic disorders and other familial traits in non-laboratory animals. Nucleic Acids Res. 2003;31(1):275–277. doi: 10.1093/nar/gkg074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill RE, Hastie ND. Accelerated evolution in the reactive centre regions of serine protease inhibitors. Nature. 1987;326(6108):96–99. doi: 10.1038/326096a0. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 18.Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA. TreeSAAP: Selection on amino acid properties using phylogenetic trees. Bioinformatics. 2003;19(5):671–672. doi: 10.1093/bioinformatics/btg043. [DOI] [PubMed] [Google Scholar]
- 19.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7(10):e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savage RJG. Evolution in carnivorous mammals. Palaeontology. 1977;20:237–271. [Google Scholar]
- 21.Heffner RS, Heffner HE. Hearing range of the domestic cat. Hear Res. 1985;19(1):85–88. doi: 10.1016/0378-5955(85)90100-5. [DOI] [PubMed] [Google Scholar]
- 22.Riazuddin S, et al. Tricellulin is a tight-junction protein necessary for hearing. Am J Hum Genet. 2006;79(6):1040–1051. doi: 10.1086/510022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su C-C, et al. Mechanism of two novel human GJC3 missense mutations in causing non-syndromic hearing loss. Cell Biochem Biophys. 2013;66(2):277–286. doi: 10.1007/s12013-012-9481-8. [DOI] [PubMed] [Google Scholar]
- 24.Huang AS, Kim LA, Fawzi AA. Clinical characteristics of a large choroideremia pedigree carrying a novel CHM mutation. Arch Ophthalmol. 2012;130(9):1184–1189. doi: 10.1001/archophthalmol.2012.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ewer RF. The Carnivores. Cornell Univ Press; New York: 1973. [Google Scholar]
- 26.Miller KA, et al. Inner ear morphology is perturbed in two novel mouse models of recessive deafness. PLoS ONE. 2012;7(12):e51284. doi: 10.1371/journal.pone.0051284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bauer JE. Metabolic basis for the essential nature of fatty acids and the unique dietary fatty acid requirements of cats. J Am Vet Med Assoc. 2006;229(11):1729–1732. doi: 10.2460/javma.229.11.1729. [DOI] [PubMed] [Google Scholar]
- 28.Butterwick RF, Salt C, Watson TDG. Effects of increases in dietary fat intake on plasma lipid and lipoprotein cholesterol concentrations and associated enzyme activities in cats. Am J Vet Res. 2012;73(1):62–67. doi: 10.2460/ajvr.73.1.62. [DOI] [PubMed] [Google Scholar]
- 29.Johansson A, et al. Identification of ACOX2 as a shared genetic risk factor for preeclampsia and cardiovascular disease. Eur J Hum Genet. 2011;19(7):796–800. doi: 10.1038/ejhg.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unno N, et al. A single nucleotide polymorphism in the plasma PAF acetylhydrolase gene and risk of atherosclerosis in Japanese patients with peripheral artery occlusive disease. J Surg Res. 2006;134(1):36–43. doi: 10.1016/j.jss.2006.02.058. [DOI] [PubMed] [Google Scholar]
- 31.Cho YS, et al. The tiger genome and comparative analysis with lion and snow leopard genomes. Nat Commun. 2013;4:2433. doi: 10.1038/ncomms3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S, et al. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell. 2014;157(4):785–794. doi: 10.1016/j.cell.2014.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han MV, Thomas GWC, Lugo-Martinez J, Hahn MW. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 2013;30(8):1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- 34.De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: A computational tool for the study of gene family evolution. Bioinformatics. 2006;22(10):1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 35.Kitchener AC. The Natural History of the Wild Cats. Cornell Univ Press; New York: 1991. [Google Scholar]
- 36.Gilad Y, Przeworski M, Lancet D, Lancet D, Pääbo S. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2004;2(1):E5. doi: 10.1371/journal.pbio.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warren WC, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453(7192):175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li G, Janecka JE, Murphy WJ. Accelerated evolution of CES7, a gene encoding a novel major urinary protein in the cat family. Mol Biol Evol. 2011;28(2):911–920. doi: 10.1093/molbev/msq281. [DOI] [PubMed] [Google Scholar]
- 39.Cameron-Beaumont C, Lowe SE, Bradshaw CJA. Evidence suggesting preadaptation to domestication throughout the small Felidae. Biol J Linn Soc Lond. 2002;75(3):361–366. [Google Scholar]
- 40.Driscoll CA, Macdonald DW, O’Brien SJ. From wild animals to domestic pets, an evolutionary view of domestication. Proc Natl Acad Sci USA. 2009;106(Suppl 1):9971–9978. doi: 10.1073/pnas.0901586106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Axelsson E, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495(7441):360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- 42.Alhaddad H, et al. Extent of linkage disequilibrium in the domestic cat, Felis silvestris catus, and its breeds. PLoS ONE. 2013;8(1):e53537. doi: 10.1371/journal.pone.0053537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukuda E, et al. Down-regulation of protocadherin-α A isoforms in mice changes contextual fear conditioning and spatial working memory. Eur J Neurosci. 2008;28(7):1362–1376. doi: 10.1111/j.1460-9568.2008.06428.x. [DOI] [PubMed] [Google Scholar]
- 44.Mead AN, Stephens DN. Selective disruption of stimulus-reward learning in glutamate receptor gria1 knock-out mice. J Neurosci. 2003;23(3):1041–1048. doi: 10.1523/JNEUROSCI.23-03-01041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mead AN, Brown G, Le Merrer J, Stephens DN. Effects of deletion of gria1 or gria2 genes encoding glutamatergic AMPA-receptor subunits on place preference conditioning in mice. Psychopharmacology (Berl) 2005;179(1):164–171. doi: 10.1007/s00213-004-2071-8. [DOI] [PubMed] [Google Scholar]
- 46.Horn KE, et al. DCC expression by neurons regulates synaptic plasticity in the adult brain. Cell Reports. 2013;3(1):173–185. doi: 10.1016/j.celrep.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 47.Yetnikoff L, Almey A, Arvanitogiannis A, Flores C. Abolition of the behavioral phenotype of adult netrin-1 receptor deficient mice by exposure to amphetamine during the juvenile period. Psychopharmacology (Berl) 2011;217(4):505–514. doi: 10.1007/s00213-011-2312-6. [DOI] [PubMed] [Google Scholar]
- 48.Takebe A, et al. Microarray analysis of PDGFRα+ populations in ES cell differentiation culture identifies genes involved in differentiation of mesoderm and mesenchyme including ARID3b that is essential for development of embryonic mesenchymal cells. Dev Biol. 2006;293(1):25–37. doi: 10.1016/j.ydbio.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 49.Demirkan A, et al. DIAGRAM Consortium CARDIoGRAM Consortium CHARGE Consortium EUROSPAN Consortium Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet. 2012;8(2):e1002490. doi: 10.1371/journal.pgen.1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkins AS, Wrangham RW, Fitch WT. The “domestication syndrome” in mammals: A unified explanation based on neural crest cell behavior and genetics. Genetics. 2014;197(3):795–808. doi: 10.1534/genetics.114.165423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kobayashi K, Jakt LM, Nishikawa SI. Epigenetic regulation of the neuroblastoma genes, Arid3b and Mycn. Oncogene. 2013;32(21):2640–2648. doi: 10.1038/onc.2012.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei Z, Yan J, Lu Q, Pan L, Zhang M. Cargo recognition mechanism of myosin X revealed by the structure of its tail MyTH4-FERM tandem in complex with the DCC P3 domain. Proc Natl Acad Sci USA. 2011;108(9):3572–3577. doi: 10.1073/pnas.1016567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu X-J, et al. Myosin X regulates netrin receptors and functions in axonal path-finding. Nat Cell Biol. 2007;9(2):184–192. doi: 10.1038/ncb1535. [DOI] [PubMed] [Google Scholar]
- 54.Hwang Y-S, Luo T, Xu Y, Sargent TD. Myosin-X is required for cranial neural crest cell migration in Xenopus laevis. Dev Dyn. 2009;238(10):2522–2529. doi: 10.1002/dvdy.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown KR, Jurisica I. Unequal evolutionary conservation of human protein interactions in interologous networks. Genome Biol. 2007;8(5):R95. doi: 10.1186/gb-2007-8-5-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hauswirth R, et al. Mutations in MITF and PAX3 cause “splashed white” and other white spotting phenotypes in horses. PLoS Genet. 2012;8(4):e1002653. doi: 10.1371/journal.pgen.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rubin CJ, et al. Strong signatures of selection in the domestic pig genome. Proc Natl Acad Sci. 2012;109(48):19529–19536. doi: 10.1073/pnas.1217149109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reissmann M, Ludwig A. Pleiotropic effects of coat colour-associated mutations in humans, mice and other mammals. Semin Cell Dev Biol. 2013;24(6-7):576–586. doi: 10.1016/j.semcdb.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 59.Menotti-Raymond M, et al. An autosomal genetic linkage map of the domestic cat, Felis silvestris catus. Genomics. 2009;93(4):305–313. doi: 10.1016/j.ygeno.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ross-Ibarra J. The evolution of recombination under domestication: A test of two hypotheses. Am Nat. 2004;163(1):105–112. doi: 10.1086/380606. [DOI] [PubMed] [Google Scholar]
- 61.Coop G, Przeworski M. An evolutionary view of human recombination. Nat Rev Genet. 2007;8(1):23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- 62.Kanan Y, Wicker LD, Al-Ubaidi MR, Mandal NA, Kasus-Jacobi A. Retinol dehydrogenases RDH11 and RDH12 in the mouse retina: Expression levels during development and regulation by oxidative stress. Invest Ophthalmol Vis Sci. 2008;49(3):1071–1078. doi: 10.1167/iovs.07-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kurushima JD, et al. Variation of cats under domestication: Genetic assignment of domestic cats to breeds and worldwide random-bred populations. Anim Genet. 2013;44(3):311–324. doi: 10.1111/age.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Anonymous (July 22, 1871). Crystal Palace - Summer concert today, Cat Show on July 13. Penny Illustrated Paper. p 16.
- 65.David VA, et al. Endogenous retrovirus insertion in the KIT oncogene determines white and white spotting in domestic cats. G3 (Bethesda) 2014 doi: 10.1534/g3.114.013425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cooper MP, Fretwell N, Bailey SJ, Lyons LA. White spotting in the domestic cat (Felis catus) maps near KIT on feline chromosome B1. Anim Genet. 2006;37(2):163–165. doi: 10.1111/j.1365-2052.2005.01389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Geissler EN, Ryan MA, Housman DE. The dominant-white spotting (W) locus of the mouse encodes the c-kit proto-oncogene. Cell. 1988;55(1):185–192. doi: 10.1016/0092-8674(88)90020-7. [DOI] [PubMed] [Google Scholar]
- 68.Blechman JM, et al. The fourth immunoglobulin domain of the stem cell factor receptor couples ligand binding to signal transduction. Cell. 1995;80(1):103–113. doi: 10.1016/0092-8674(95)90455-7. [DOI] [PubMed] [Google Scholar]
- 69.Miller JR, et al. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics. 2008;24(24):2818–2824. doi: 10.1093/bioinformatics/btn548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flicek P, et al. Ensembl 2012. Nucleic Acids Res. 2012;40(database issue):D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katoh K, Toh H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 2010;26(15):1899–1900. doi: 10.1093/bioinformatics/btq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013. Nucleic Acids Res. 2013;41(web server issue):W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ashburner M, et al. The Gene Ontology Consortium Gene Ontology: Tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cerami EG, et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39(database issue):D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kelder T, et al. WikiPathways: Building research communities on biological pathways. Nucleic Acids Res. 2012;40(database issue):D1301–D1307. doi: 10.1093/nar/gkr1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(database issue):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 78.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koboldt DC, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kofler R, Pandey RV, Schlötterer C. PoPoolation2: Identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq) Bioinformatics. 2011;27(24):3435–3436. doi: 10.1093/bioinformatics/btr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rubin C-J, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464(7288):587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- 82.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Untergasser A, et al. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.