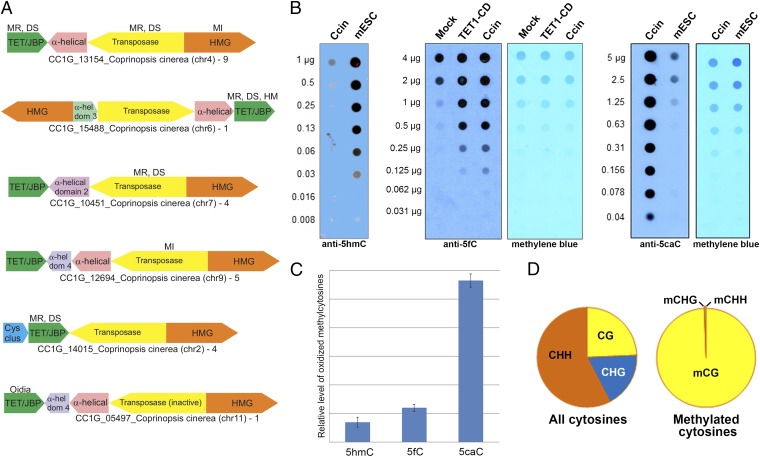
Fig. 1.
TET/JBP-coding transposons and oxi-mCs. (A) Genomic organization of the Kyakuja transposons in C. cinerea. Genes are depicted as arrows pointing from the 5′ to the 3′ direction of the coding sequence. Gene neighborhoods are labeled with the name of the TET/JBP gene, species name, and the sequence identification (gi) number with the chromosome number in brackets. The number of complete elements with a particular organization in the genome is shown next to the label. Five of the depicted transposons represent the more prevalent Kyakuja-1 element, which contains a gene coding for a predominantly α-helical protein between the TET/JBP and transposase genes that may also be fused to the transposase; the sixth is a less frequent Kyakuja-2 element that contains a gene for a cysteine-rich protein with conserved cysteine residues (Cys-clus) 5′ to the TET/JBP gene. In most Kyakuja elements, the TET/JBP gene is encoded on the opposite strand of DNA relative to the gene for the transposase, with the two in a head-to-head orientation; the TET/JBP gene and the gene encoding the HMG domain-containing protein (HMG) are usually on the same strand. The GenBank entry for CC1G_15488 and the transposase associated with CC1G_05497 fuses several distinct domains on the same strand. These are depicted as separate genes. α-hel dom, α-helical domain; DS, dikaryon-specific; HM, expressed in haploid mycelium; MI, meiotically induced; MR, meiotically repressed; MS, meiosis-specific; oidia, expressed in oidia. Stage-specific gene expression information is from Burns et al. (50). (B) Dot blots showing the relative abundance of 5hmC (Left), 5fC (Middle), and 5caC (Right) in C. cinerea (Ccin) compared with mouse embryonic stem cell DNA. TET1CD, DNA from HEK293 cells transfected with the TET1 catalytic domain. (C) Relative levels of 5hmC, 5fC, and 5caC in C. cinerea estimated by MS. (D) All ∼18.5 million cytosines (on both strands) in the 13 assembled chromosomes of the C. cinerea genome were categorized into three groups according to their sequence context (H = A, C, or T). Although only one fourth of all cytosines occur in the CpG context, more than 99% of all methylated cytosines (or hydroxymethylated cytosines, as bisulfite sequencing cannot distinguish between 5mC and 5hmC) are observed at CpGs.