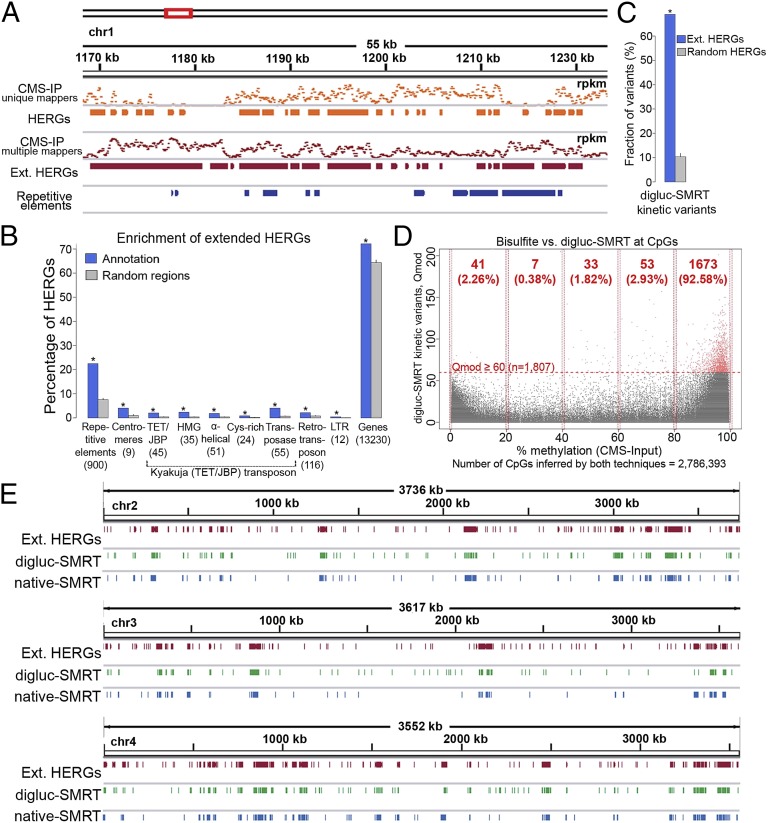
Fig. 3.
Mapping oxi-mCs in the C. cinerea genome. (A) Anti-CMS IP coverage [in reads per kilobase per million (rpkm)] of short reads mapping to only one position in the genome (unique mappers) or as many as 100 positions in the genome (multiple mappers). By considering only uniquely mapped reads, a substantial number of repetitive genomic regions cannot be covered by input or CMS-IP data. To address this problem, we also considered reads that mapped to as many as 100 different genomic regions in input and CMS-IP analyses (SI Materials and Methods). Extended HERGs are obtained by merging HERGs identified by the two alternative approaches. (B) To estimate the enrichment of HERGs at different annotated genomic regions, we performed 100 random samplings of the same number of genomic regions with the same size distribution (SI Materials and Methods). This analysis showed substantial enrichment of HERGs at repetitive elements and Kyakuja transposons (P ≤ 0.001, empirical test). A substantial amount of HERGs (72%) accumulate at genes and 4% of HERGs mapped to annotated centromeres. (C) An analogous analysis shows substantial enrichment of digluc-SMRT kinetic variants at extended HERGs (P ≤ 0.001), suggesting a high agreement between methods. (D) Single base-level comparison of cytosines in a CpG context covered by bisulfite (CMS input, x axis) and digluc-SMRT (y axis) sequencing (n = 2,786,393). Horizontal dashed red line represents a Qmod 60 threshold applied for defining digluc-SMRT kinetic variants. Bisulfite methylation shows a common bimodal distribution, and 5hmC is detected by digluc-SMRT essentially only in the high range of bisulfite methylation that is incorrectly considered as highly methylated by bisulfite sequencing. (E) Chromosome-wide views of extended HERGs (CMS IP), digluc-SMRT, and native SMRT sequencing data shows high correspondence among the distributions of oxi-mC species detected by these different technologies.