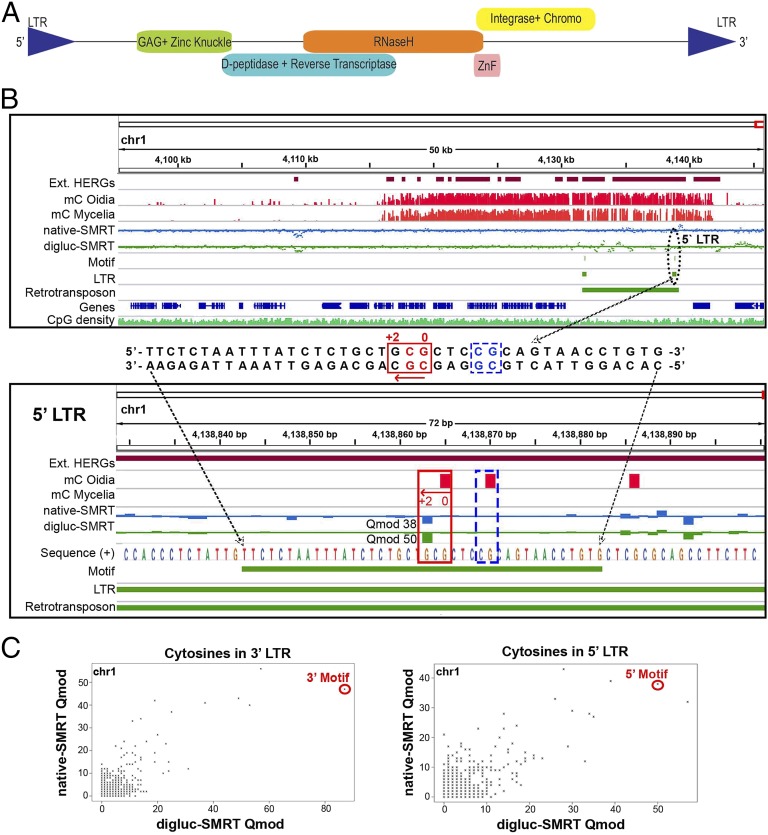
Fig. 5.
oxi-mCs identify LTRs of Ty/gypsy-like retrotransposons. (A) Schematic view of a complete retrotransposon distantly related to the Ty-like elements of ascomycete fungi, encoding a polyprotein with serial GAG, Zn-Knuckle, aspartyl peptidase, reverse transcriptase, RNase H, ZnF, integrase, and chromodomain. (B) Regional view (Top) of a complete copy of the retrotransposon showing its location in a genomic region free of annotated genes but with high abundance of 5mC toward the end of chromosome 1. A conserved DNA sequence (Middle) has been identified in the LTRs of the retrotransposons as a result of a single modified cytosine as highlighted in a local view (Bottom) of the motif in the 5′ LTR of the retrotransposon shown above. The presence of elevated signals in native-SMRT (strand specific blue track, Qmod value 38) and digluc-SMRT (strand-specific green track, Qmod value 50) sequencing, suggests that the cytosine in the CpG context located two bases upstream of the actual signal at the minus strand reflects a population of 5hmC and either 5fC or 5caC. Units are non–strand-specific fractions (as percentages) of the sum of 5mC and 5hmC compared with all sequenced cytosines at each position deduced by bisulfite sequencing for mC oidia and mC mycelia, and strand-specific Qmod values for digluc-SMRT and native SMRT sequencing. (C) Direct correlation of digluc-SMRT (x axis) and native SMRT (y axis) sequencing-derived Qmod values of all cytosines in the 5′ (Top) and 3′ (Bottom) LTR of the retrotransposon shown in Fig. 5B. Highlighted by red circles are the single modified cytosines in the LTR motif.