Fig. 1.
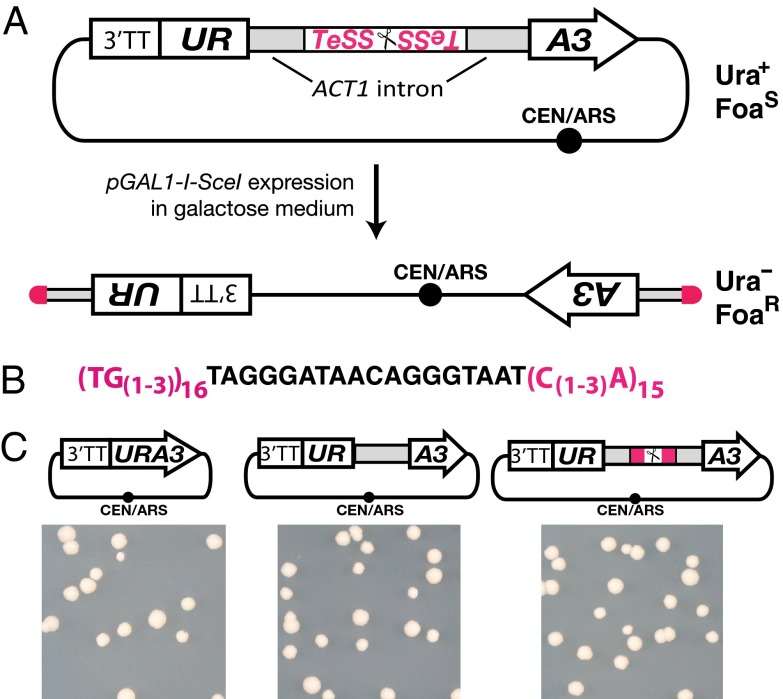
Telomerator design and function. (A) The telomerator encodes a selectable URA3 gene sequence interrupted by an intron (ACT1 intron) that harbors a homing endonuclease recognition site (depicted by scissors). The recognition site is flanked by telomere seed sequences (TeSS). The URA3 promoter sequence also encodes a strong 3′ transcriptional terminator sequence (3′TT). The telomerator cassette is encoded on a circular DNA molecule yielding cells that grow on medium lacking uracil (Ura+) but are sensitive to the uracil counterselection drug Foa (FoaS), assuming the circular DNA molecule encodes at least one essential genetic part. Expression of I-SceI, under the control of the GAL1 promoter (pGAL1), is induced by growth in galactose medium. Cells harboring newly linearized molecules should no longer grow on plates lacking uracil (Ura–) and should be resistant to Foa (FoaR). (B) Detailed sequence information for the TeSSs (pink) flanking the I-SceI recognition sequence (black) of the telomerator. (C) The telomerator complements yeast cell growth on medium lacking uracil. Plasmids encoding a native URA3 gene (Left), a URA3 gene interrupted by the intron of ACT1 (Middle), or a URA3 gene interrupted by a telomerator sequence encoded within the ACT1 intron (Right) were transformed into a yeast strain (BY4741) incapable of growing on medium lacking uracil (SC–Ura). Growth of the resulting transformed colonies on SC–Ura was indistinguishable, indicating normal expression of the URA3 gene product from the telomerator-encoding construct.