Abstract
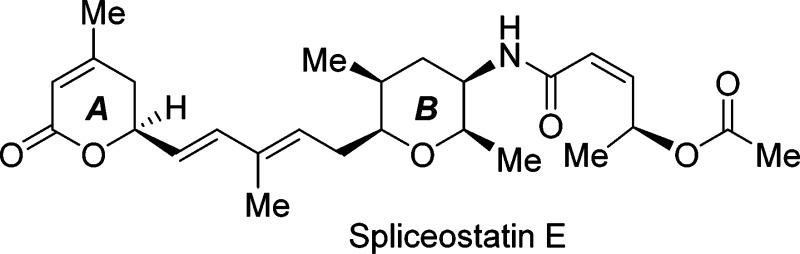
An enantioselective total synthesis of spliceostatin E has been accomplished. The δ-lactone unit A was constructed from readily available (R)-glycidyl alcohol using a ring-closing olefin metathesis as the key reaction. A cross-metathesis of ring A containing δ-lactone and the functionalized tetrahydropyran B-ring provided spliceostatin E. Our biological evaluation of synthetic spliceostatin E revealed that it does not inhibit splicing in vitro and does not impact speckle morphology in cells. Spliceostatin E was reported to possess potent antitumor activity.
Splicing is an essential step in gene expression in which noncoding sequences (introns) are removed from RNA transcripts and coding sequences (exons) are ligated to form messenger RNA (mRNAs).1 A complex ribonucleoprotein machinery, known as spliceosome, facilitates this splicing event.2 The splicing levels are generally increased in cancer cells, and as a consequence, spliceosome inhibitors have been implicated in anticancer drug development.3−5 Pladienolide B, 1 (Figure 1), and their derivatives have been shown to inhibit spliceosome by binding to the SF3b subunit of spliceosome.6 A semisynthetic pladienolide B derivative has entered in human clinical trials.7 Structurally distinct, FR901464, 2, and its methylated derivative, spliceostatin A, 3, have also been shown to inhibit in vitro splicing by binding to the SF3b subunit of the spliceosome.8 FR901464, 2, was isolated by Nakajima and co-workers at the Fujisawa Pharmaceutical Co. from the fermentation broth of strain no. 2663 in 1996.9,10 It exhibited potent anticancer activity with IC50 values ranging from 0.6 to 3.4 nM against numerous human cancer cell lines. Yoshida and co-workers then reported spliceostatin A with similar activity as FR901464.11,12 Not surprisingly, both FR901464 and spliceostatin A have attracted considerable synthetic interest.13−15 We recently reported the synthesis and structure–activity studies of FR901464 and spliceostatin A.16,17 FR901464 and spliceostatin A are structurally quite complex and thus posed a challenge as a starting point for structural modification.
Figure 1.
Structures of pladienolide B, FR901464, and spliceostatins A and E.
Very recently, He and co-workers reported the isolation of a number of spliceostatin class natural products from the fermentation broth of FERM BP3421, which was later renamed as Burkholderia sp.18 Among these new spliceostatins, structurally less complex spliceostatin E, 3, exhibited good potency against multiple human cancer cell lines with IC50 values ranging from 1.5 to 4.1 nM. The structure of spliceostatin E was elucidated by extensive spectroscopic studies.18 Spliceostatin E structure contains a 5, 6-dihydro-α-pyrone unit (A ring) and a common highly functionalized B ring. The structural features of spliceostatin E appear to be very attractive for structural variation and optimization of potency. Herein, we report an enantioselective synthesis of spliceostatin E. We have also evaluated its spliceosome inhibitory activity and effect on cells.
Our synthetic strategy for spliceostatin E is shown in Scheme 1. We planned to carry out a cross-metathesis of vinyl dihydropyranone 5 and tetrahydropyranyl diene 6 to construct spliceostatin E. Optically active 5,6-dihydro-α-pyrone ring A would be synthesized from (S)-glycidyl silyl ether 7, which can be readily prepared from commercially available (R)-glycidyl alcohol. The tetrahydropyranyl ring B fragment would be obtained by formation of an amide between previously prepared amine 8(16,17) and optically active acid 9.
Scheme 1. Retrosynthesis of Spliceostatin E.
The synthesis of (S)-dihydropyranone 5 is shown in Scheme 2. Opening of the epoxide 7 with isopropenylmagnesium bromide in the presence of a catalytic amount of CuCN (10 mol %) at −20 to +23 °C for 30 min provided the corresponding homoallylic alcohol.19 Reaction of the resulting alcohol with acrolein diethyl acetal in the presence of a catalytic amount of PPTS in benzene at 23 °C for 12 h afforded ethyl acetal 10 as a mixture (1:1 by 1H NMR) of diastereomers in 74% yield. This mixture was subjected to ring-closing metathesis using Grubbs’ second-generation catalyst20,21 in the presence of NaHCO3 in CH2Cl2 at reflux. This condition resulted in a 1:1 mixture of dihydropyran derivative in 86% yield. These isomers could not be separated by silica gel chromatography. However, treatment of this mixture with a catalytic amount of camphorsulfonic acid (CSA) (10 mol %) in ethanol for 2 h provided 11 as a major diastereomer (ratio >20:1). Exposure of 11 to nBu4N+F– in THF for 3 h at 23 °C provided the corresponding alcohol in near quantitative yield. Swern oxidation of the alcohol afforded the aldehyde which was subjected to Wittig olefination at 0 °C to furnish olefin 13 in 54% yield over three steps. Oxidation of ethyl acetal 13 with PCC in CH2Cl222 at 23 °C for 5 h afforded optically active dihydropyranone derivative 5 in 73% yield.
Scheme 2. Synthesis of (S)-Dihydropyranone, 5.
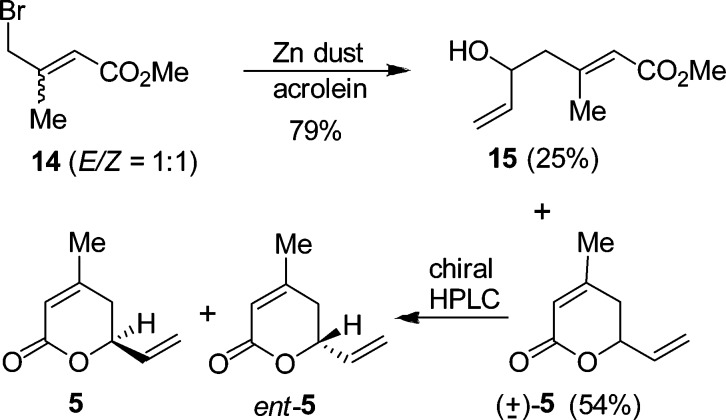
To provide rapid access to (S)-dihydropyranone ent-5, we have explored an alternative route. As shown in Scheme 3, allyl bromide 14 was synthesized by bromination of 3,3-dimethyl acrylate with N-bromosuccinimide as described by Fallis and Lei.23 The E/Z mixture (1:1) was exposed to vinylogous Reformatsky reaction with acrolein to provide allylic alcohol 15 and racemic dihydropyranone (±)-5.24 These products were separated by silica gel chromatography (54% yield, 1:1 mixture). The racemic mixture was separated by chiral HPLC using a Chiralpak IA column to provide access to both (R)- and (S)-dihydropyranone derivatives.25
Scheme 3. Synthesis of Racemic Dihyropyranone.
The synthesis of Z-allylic acetate 9 is outlined in Scheme 4. Commercially available (S)-3-butyn-2-ol was protected as TES-ether with TESOTf and Et3N in CH2Cl2. Treatment of this TES-ether with nBuLi followed by reaction with ethyl chloroformate afforded alkylated product 17 in 81% yield.26 Removal of the TES group followed by saponification of the ethyl ester with aqueous LiOH provided the corresponding acid. Acylation of this acid with acetyl chloride at 23 °C for 5 h furnished acetate 18. Catalytic hydrogenation of acetate 18 over Lindlar’s catalyst as described by us previously16,17 provided optically active Z-alkene acetate.
Scheme 4. Synthesis of Z-Alkene Acid 9.
The synthesis of spliceostatin E is shown in Scheme 5. Amide coupling of acid 9 and amine 8, which was prepared as described by us previously, provided amide derivative 6 in 60% yield. Cross-metathesis of diene 6 and alkene 5 with Grubbs’ second-generation catalyst in CH2Cl2 at reflux afforded spliceostatin E, 4, in 71% yield. The 1H and 13C NMR of our synthetic spliceostatin E [[α]23D −70 (c 0.2, CHCl3)] are in full agreement with the reported spectra of natural spliceostatin E.18
Scheme 5. Synthesis of Spliceostatin E.
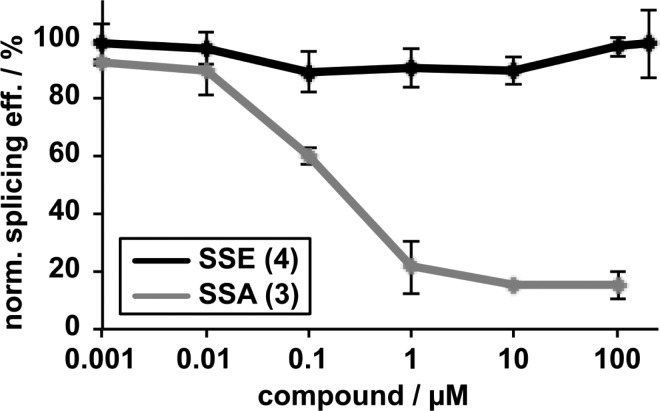
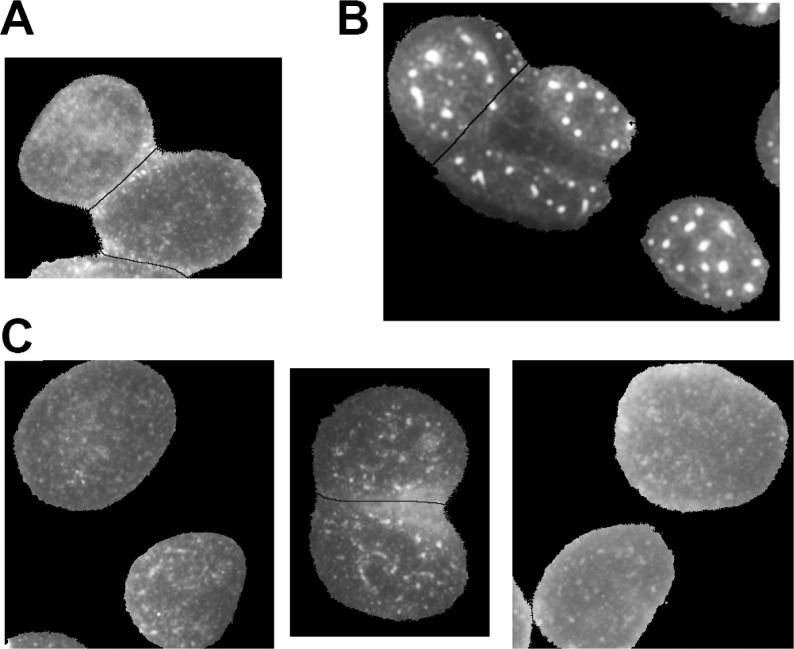
The biological properties of synthetic spliceostatin E, 4, were evaluated (Figure 2) in an in vitro splicing system as previously described.27 Strikingly, the compound showed no inhibition of splicing in this system, even at 200 μM concentration. In contrast, spliceostatin A in the same assay shows strong splicing inhibition. This result was counter to our expectations because of the recent demonstration of comparable cytotoxicity for both compounds.18 Therefore, we examined the effect of spliceostatin E on the structure of nuclear speckles in HeLa cells (Figure 3). Potent splicing inhibitors including pladienolide B and spliceostatin A12 cause speckles to increase in size. We see no strong difference between cells treated with DMSO and spliceostatin E, which correlates with the in vitro splicing results.
Figure 2.
Impact of analogues on in vitro splicing. Average splicing efficiency vs inhibitor concentration normalized to DMSO control. SSE, spliceostatin E; SSA, spliceostatin A.
Figure 3.
Changes in nuclear speckle morphology. SFRS2 (SC35) immunofluorescence in HeLa cells nuclei treated with (A) DMSO, (B) 1 μM pladienolide B, and (C) 0.1, 1, and 10 μM spliceostatin E, left to right.
In summary, we have accomplished the first enantioselective synthesis of reported spliceostatin E (the optical rotation was not reported for spliceostatin E). Enantioselective synthesis of dihydropyranone ring A was achieved from commercially available (R)-glycidyl alcohol using a ring-closing olefin metathesis as the key reaction. A Reformatsky reaction of methyl 3-bromomethyl crotonate with acrolein also provided rapid access to racemic dihydropyranone which was separated by chiral HPLC. A cross-metathesis of two fragments using Grubbs catalyst efficiently provided the final product. The synthesis is convergent and amenable to the synthesis of structural variants. We have also evaluated spliceosome inhibitory activity of spliceostatin E and compared its activity with spliceostatin A. Surprisingly, spliceostatin E does not inhibit splicing in vitro and does not impact speckle morphology in cells. The design and synthesis of structural variants of spliceostatins are in progress. These analogs will be important to clarify the link between splicing inhibition and changes in cellular function induced by these remarkable compounds.
Acknowledgments
Financial support from the National Institutes of Health and Purdue University is gratefully acknowledged.
Supporting Information Available
General experimental procedures and characterization data for all new products. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Jurica M. S.; Roybal G. A. Ency. Biol. Chem. 2013, 4, 167–171. [Google Scholar]
- Will C. L.; Luhrmann R. Cold Spring Harbor Perspect Biol. 2011, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Alphen R. J.; Wiemer E. A.; Burger H.; Eskens F. A. Br. J. Cancer 2009, 100, 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal S.; Vigevani L; Valcárcel J. Nat. Rev. Drug Discov. 2012, 11, 847–859. [DOI] [PubMed] [Google Scholar]
- Shkreta L.; Bell B.; Revil T.; Venables J. P.; Prinos P.; Elela S. A.; Chabot B. Cancer Treat. Res. 2013, 158, 41–94. [DOI] [PubMed] [Google Scholar]
- Kotake Y.; Sagane K.; Owa T.; Mimori-Kiyosue Y.; Shimizu H.; Uesugi M.; Ishihama Y.; Iwata M.; Mizui Y. Nat. Chem. Biol. 2007, 3, 570–575. [DOI] [PubMed] [Google Scholar]
- Eskens F. A. L. M.; Ramos F. J.; Burger H.; O’Brien J. P.; Piera A.; de Jonge M. J. A.; Mizui Y.; Wiemer E. A. C.; Carreras M. J.; Baselga J.; Tabernero J. Clin. Cancer Res. 2013, 19, 6296–6304. [DOI] [PubMed] [Google Scholar]
- Kaida D.; Motoyoshi H.; Tashiro E.; Nojima T.; Hagiwara M.; Ishigami K.; Watanabe H.; Kitahara T.; Yoshida T.; Nakajima H.; Tani T.; Horinouchi S.; Yoshida M. Nat. Chem. Biol. 2007, 3, 576–583. [DOI] [PubMed] [Google Scholar]
- Nakajima H.; Sato B.; Fujita T.; Takase S.; Terano H.; Okuhara M. J. Antibiot. 1996, 49, 1196–1203. [DOI] [PubMed] [Google Scholar]
- Nakajima H.; Hori Y.; Terano H.; Okuhara M.; Manda T.; Matsumoto S.; Shimomura K. J. Antibiot. 1996, 49, 1204–1211. [DOI] [PubMed] [Google Scholar]
- Motoyoshi H.; Horigome M.; Ishigami K.; Yoshida T.; Horinouchi S.; Yoshida M.; Watanabe H.; Kitahara T. Biosci. Biotechnol. Biochem. 2004, 68, 2178–2182. [DOI] [PubMed] [Google Scholar]
- Kaida D.; Motoyoshi H.; Tashiro E.; Nojima T.; Hagiwara M.; Ishigami K.; Watanabe H.; Kitahara T.; Yoshida T.; Nakajima H.; Tani T.; Horinouchi S.; Yoshida M. Nat. Chem. Biol. 2007, 3, 576–583. [DOI] [PubMed] [Google Scholar]
- a Thompson C. F.; Jamison T. F.; Jacobsen E. N. J. Am. Chem. Soc. 2000, 122, 10482–10483. [Google Scholar]; b Thompson C. F.; Jamison T. F.; Jacobsen E. N. J. Am. Chem. Soc. 2001, 123, 9974–9983. [DOI] [PubMed] [Google Scholar]
- a Horigome M.; Motoyoshi H.; Watanabe H.; Kitahara T. Tetrahedron Lett. 2001, 42, 8207–8210. [Google Scholar]; b Motoyoshi H.; Horigome M.; Watanabe H.; Kitahara T. Tetrahedron 2006, 62, 1378–1389. [Google Scholar]
- a Albert B. J.; Koide K. Org. Lett. 2004, 6, 3655–3658. [DOI] [PubMed] [Google Scholar]; b Albert B. J.; Sivaramakrishnan A.; Naka T.; Koide K. J. Am. Chem. Soc. 2006, 128, 2792–2793. [DOI] [PubMed] [Google Scholar]; c Albert B. J.; Sivaramakrishnan A.; Naka T.; Czaicki N. L.; Koide K. J. Am. Chem. Soc. 2007, 129, 2648–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. K.; Chen Z. H. Org. Lett. 2013, 15, 5088–5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. K.; Chen Z. H.; Effenberger K. A.; Jurica M. S. J. Org. Chem. 2014, 79, 5697–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H.; Ratnayake A. S.; Janso J. E.; He M.; Yang H. Y.; Loganzo F.; Shor B.; O’Donnell C. J.; Koehn F. E. J. Nat. Prod. 2014, 77, 1864–1870. [DOI] [PubMed] [Google Scholar]
- Ghosh A. K.; Wang Y.; Kim J. T. J. Org. Chem. 2001, 66, 8973–8982. [DOI] [PubMed] [Google Scholar]
- Chatterjee A. K.; Choi T. L.; Sanders D. P.; Grubbs R. H. J. Am. Chem. Soc. 2003, 125, 11360–11370. [DOI] [PubMed] [Google Scholar]
- Pruent J. Curr. Top. Med. Chem. 2005, 5, 1559–1577. [DOI] [PubMed] [Google Scholar]
- Jayasinghe S.; Venukadasula P. K.; Hanson P. R. Org. Lett. 2014, 16, 122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei B.; Fallis A. G. Can. J. Chem. 1991, 69, 1450–1456. [Google Scholar]
- Ghosh A. K.; Cheng X. Org. Lett. 2011, 13, 4108–4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- For details, see the Supporting Information.
- Crimmins M. T.; Choy A. L. J. Am. Chem. Soc. 1997, 119, 10237–10238. [Google Scholar]
- Effenberger K. A.; Anderson D. D.; Bray W. M.; Prichard B. E.; Ma N.; Adams M. S.; Ghosh A. K.; Jurica M. S. J. Biol. Chem. 2014, 289, 1938–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.