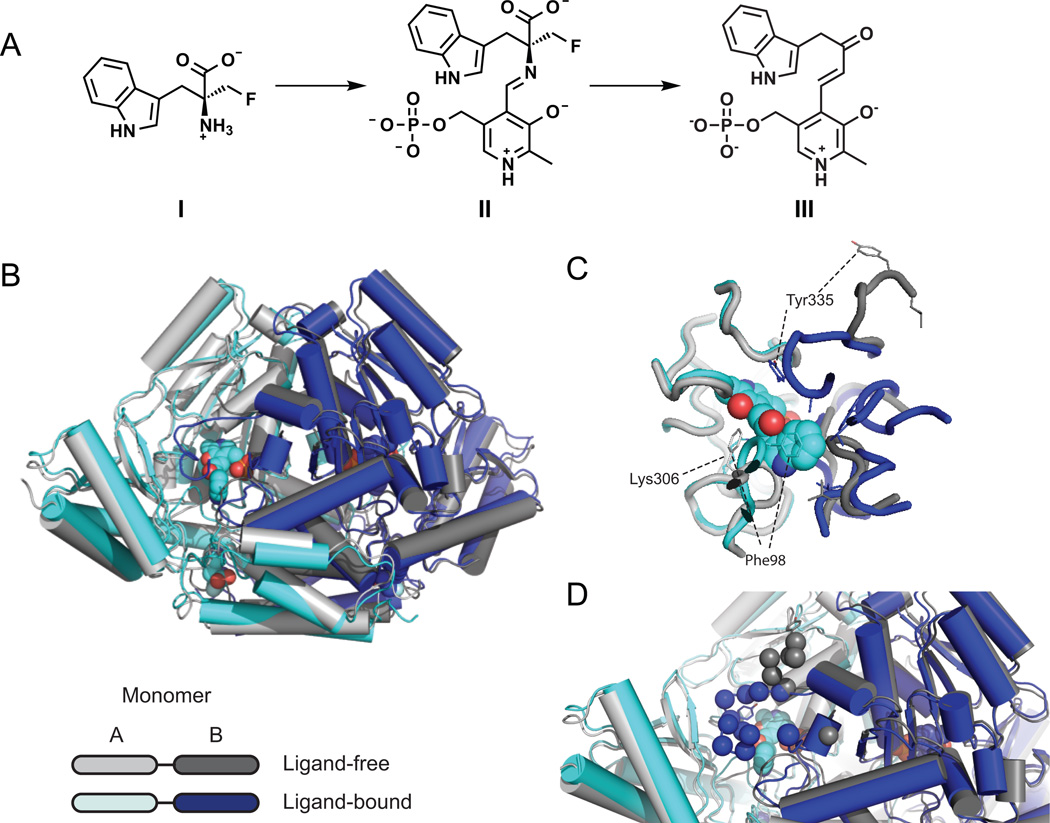
Figure 3. Crystal structure of apo and ligand-bound RUMGNA_01526.
(A) Schematic of proposed inhibitor mechanism: (S)-α-FMT (I) is converted to a PLP-(S)-α-FMT external aldimine intermediate (II), which is decarboxylated to a PLP-(S)-α-FMT Schiff base adduct (III). (B) Overlay of ligand-free (monomer A, light gray and monomer B, dark gray) and ligand-bound (monomer A, cyan and monomer B, blue) structures. In the active and allosteric sites, PLP-(S)-α-FMT and (S)-α-FMT (respectively) are shown in spheres. (C) Active site with PLP-(S)-α-FMT bound reveals a repositioning of Tyr335 and Phe98. In the ligand-bound structure, Lys306 is no longer covalently bound to PLP. (D) Upon engagement of (S)-α-FMT, residues 337–349 (dark blue spheres) fold over the active site, excluding solvent and forming critical interactions with the inhibitor. Dark gray spheres represent only ordered residues in apo structure. See also Figure S4 and S6.