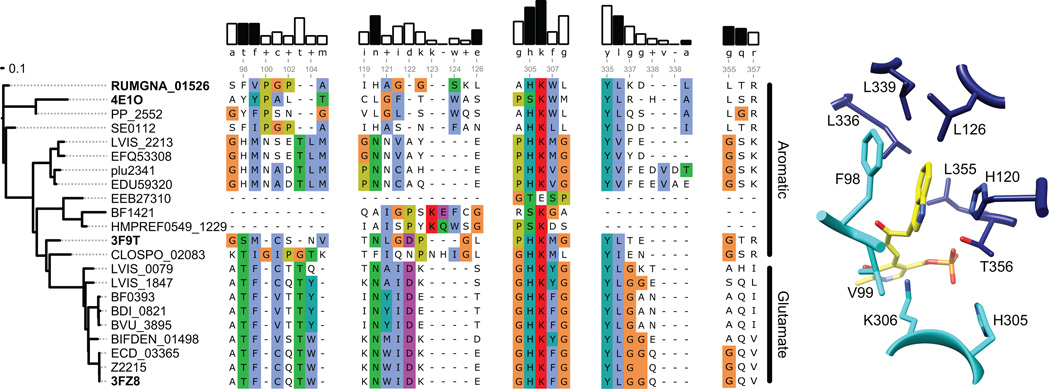
Figure 5. Sequence and structural analysis of aromatic amino acid decarboxylases.
(A) The dendrogram on the left shows the degree of sequence similarity between various decarboxylases. (B) Alignment of select amino acid decarboxylases are numbered according to the RUMGNA_01526 sequence. Four structural components of RUMGNA_01526 important for substrate binding are highlighted. The bars above consensus sequence show the degree of sequence conservation; residues from the RUMGNA_01526 structure that interact (black bars) or do not interact (white) with the tryptophan substrate are indicated. Residues in the sequence alignment are colored according to the Clustal color code (http://ekhidna.biocenter.helsinki.fi/pfam2/clustal_colours). (C) RUMGNA_10526 active site showing residues represented by black bars in (B).