Abstract
New World monkeys exhibit prominent colour vision variation due to allelic polymorphism of the long-to-middle wavelength (L/M) opsin gene. The known spectral variation of L/M opsins in primates is broadly determined by amino acid composition at three sites: 180, 277 and 285 (the ‘three-sites’ rule). However, two L/M opsin alleles found in the black-handed spider monkeys (Ateles geoffroyi) are known exceptions, presumably due to novel mutations. The spectral separation of the two L/M photopigments is 1.5 times greater than expected based on the ‘three-sites’ rule. Yet the consequence of this for the visual ecology of the species is unknown, as is the evolutionary mechanism by which spectral shift was achieved. In this study, we first examine L/M opsins of two other Atelinae species, the long-haired spider monkeys (A. belzebuth) and the common woolly monkeys (Lagothrix lagotricha). By a series of site-directed mutagenesis, we show that a mutation Y213D (tyrosine to aspartic acid at site 213) in the ancestral opsin of the two alleles enabled N294K, which occurred in one allele of the ateline ancestor and increased the spectral separation between the two alleles. Second, by modelling the chromaticity of dietary fruits and background leaves in a natural habitat of spider monkeys, we demonstrate that chromatic discrimination of fruit from leaves is significantly enhanced by these mutations. This evolutionary renovation of L/M opsin polymorphism in atelines illustrates a previously unappreciated dynamism of opsin genes in shaping primate colour vision.
Keywords: spectral differentiation, spider monkeys, visual pigments, woolly monkeys
Introduction
Colour vision polymorphism, leading to a mixed population of dichromats and trichromats, has been documented in most species of platyrrhine primates (New World monkeys) (Jacobs 2007). The presence of these diverse phenotypes is due to allelic variation of a single L/M opsin gene on the X chromosome (Mollon et al. 1984; Kawamura et al. 2001). In combination with the autosomal short wavelength (S) opsin, trichromacy is realized in females heterozygous for the L/M opsin. Dichromacy occurs in all males and homozygous females. Exceptions to this pattern have only been found in two genera, Aotus (owl monkeys) and Alouatta (howler monkeys), the former being monochromatic and nocturnal, having only an M opsin allele and lacking a functional S opsin (Jacobs et al. 1993; Hiramatsu et al. 2004; Levenson et al. 2007), and the latter considered to be routinely trichromatic, having the L and M opsin genes juxtaposed by gene duplication on the X chromosome (Jacobs et al. 1996) as in catarrhine primates (humans, apes and Old World monkeys). The exceptional intra- and interspecific variation of colour vision among New World monkeys provides a desirable platform to study the utility of colour vision to primates and to understand the evolutionary forces behind it (Mollon et al. 1984; Caine 2002; Kawamura et al. 2012).
The wavelength of maximal absorption (λmax) of the M/LWS type of vertebrate opsins (Yokoyama 2000), to which the primate L/M opsins belong, can be predicted from the amino acid composition at five sites: 180, 197, 277, 285 and 308. This is known as the ‘five-sites’ rule (Yokoyama & Radlwimmer 1998, 1999, 2001; Yokoyama et al. 2008). Among primate L/M opsins, however, the residues 197 and 308 (histidine and alanine, respectively) do not vary and the ‘five-sites’ rule can be reduced to the ‘three-sites’ rule in practice (Hiramatsu et al. 2004). The λmax of the L/M opsins with serine, tyrosine and threonine at residues 180, 277 and 285, respectively (denoted SYT), are expected to be approximately 560 nm (Yokoyama et al. 2008). The λmax values of L/M opsins with other three-site combinations can be predicted by subtracting 5, 10 and 17 nm from 560 nm in the case of alanine, phenylalanine and threonine at residues 180, 277 and 285, respectively (Yokoyama et al. 2008). In addition, interactions between these mutations are estimated to be −2 nm for S180A/T285A, +1 nm for Y277F/T285A and +4 nm for S180A/Y277F/T285A (Yokoyama et al. 2008).
We previously discovered two L/M opsin alleles, SYT and SFT, from the black-handed spider monkeys (Ateles geoffroyi), a species of New World monkeys, through field survey of a natural population in Costa Rica (Hiramatsu et al. 2005). Including them, the L/M opsins of New World monkeys can be distinguished into six types on the basis of their three-site composition: SYT, AYT, SFT, AFT, AYA and AFA (Table 1). Among primate L/M opsins, deviation of observed λmax values from that expected based on the ‘three-sites’ rule has been at most 3 nm (Yokoyama et al. 2008). In this regard, the SYT and SFT alleles of the spider monkey are an unusual exception; their deviation is much larger than expected: 7 nm in SYT (λmax 553 instead of 560 nm) and 12 nm in SFT (538 instead of 550 nm) (Table 1) (Hiramatsu et al. 2008). This deviation enlarges the spectral separation between the two resultant photopigments from the 10 nm expected to an observed 15 nm. The expected photopigment set would result in female trichromats comparable to deuteranomalous human trichomats (trichromatic colour vision based on S, L and an anomalous L-like photoreceptor), who are severely impaired in red-green chromatic discrimination (Deeb 2006). On the other hand, the observed λmax set would result in female trichromats comparable to those seen in Cebinae carrying an intermediate-λmax allele (Table 1), who are successful in discriminating stimuli using Ishihara pseudo-isochromatic plates (Saito et al. 2005). Thus, spider monkeys are predicted to have better colour vision due to their unique L/M opsins.
Table 1.
Phylogenetic distribution and expected and observed λmax values of L/M opsin types in New World monkeys
| Subfamilies | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Three-sites composition | CA* | AO† | CE‡ | PI§ | AL¶ | AT** | Expected λmax (nm) | Observed λmax (nm) | ERG λmax (nm)††† |
| SYT | ✓ | ✓ | ✓ | ✓ | ✓ | 560 | 561††,‡‡, 558§§, 553¶¶ | ∼562 | |
| AYT | ✓ | 555 | 553†† | ∼556 | |||||
| SFT | ✓ | 550 | 538¶¶ | ∼550 | |||||
| AFT | ✓ | ✓ | 545 | 545§§, 543‡‡ | ∼550 | ||||
| AYA | ✓ | ✓ | 536 | 539††,*** | ∼543 | ||||
| AFA | ✓ | ✓ | ✓ | (✓) | 532 | 532‡‡,§§ | ∼535 | ||
New World monkeys are comprised of three Families, the Cebidae (Cebinae, Callitrichinae and Aotinae), Atelidae (Atelinae and Alouattinae) and Pitheciidae (Pitheciinae and Callicebinae) (Wildman et al. 2009). Species thus far studied are
Callitrichinae: marmosets (Callithrix jacchus, C. geoffroyi), pygmy marmoset (Cebuella pygmaea), tamarins (Saguinus mystax, S. labiatus, S. fuscicollis, S. midas, S. imperator, S. geoffroyi, S. oedipus, S. bicolor), lion tamarin (Leontopithecus chrysomelas, L. chrysopygus, L. rosalia) and goeldi’ monkey (Callimico goeldii) (Shyue et al. 1998; Kawamura et al. 2001; Surridge & Mundy 2002; Surridge et al. 2005).
Aotinae: owl monkeys (Aotus lemurinus, A. azarae) (Kawamura et al. 2002; Nagao et al. 2005).
Cebinae: capuchin monkeys (Cebus olivaceus (or nigrivittatus), C. apella, C. capucinus) and squirrel monkeys (Saimiri sciureus, S. boliviensis, S. oerstedii) (Shyue et al. 1998; Cropp et al. 2002; Hiramatsu et al. 2005; Saito et al. 2005; Hiwatashi et al. 2010).
Pitheciidae: saki monkey (Pithecia irrorata) (Boissinot et al. 1998) and titi monkey (Callicebus brunneus) (Bunce et al. 2011).
Alouattinae: howler monkeys (Alouatta caraya, A. seniculus)] (Jacobs et al. 1996).
Atelinae: spider monkey (Ateles geoffroyi) (Hiramatsu et al. 2005; Hiwatashi et al. 2010) and muriquis (Brachyteles arachnoides, B. hypoxanthus)] (Talebi et al. 2006). The AFA is parenthesized because this is found only in a species of muriquis.
Common marmoset (C. jacchus) (Kawamura et al. 2001).
White-faced capuchin monkey (C. capucinus) (Hiramatsu et al. 2005).
Common squirrel monkey (S. sciureus) (Hiramatsu et al. 2004).
Black-handed spider monkey (A. geoffroyi) (Hiramatsu et al. 2008), boldfaced to highlight the deviation from the ‘three-sites’ expectation.
Owl monkey (A. azarae) (Hiramatsu et al. 2004; Nagao et al. 2005).
Estimates by electroretinogram (ERG) method. A widely used representative value is listed for each opsin type (Jacobs 2008).
It is yet unknown whether the L/M opsin alleles, with their unique spectral properties, found in black-handed spider monkeys are also present in other ateline species. Spider monkeys (Genus Ateles) belong to the Subfamily Atelinae with woolly moneys (Lagothrix) and muriquis (Brachyteles) with the cladistic relationship [Ateles (Lagothrix and Brachyteles)] (Wildman et al. 2009). The partial DNA sequences of the SYT and SFT alleles are reported for muriquis (Talebi et al. 2006). However, these pigments have not been reconstituted and measured in vitro, and no information on the opsin DNA sequences is yet available for other species of spider monkeys or woolly monkeys. Determining how widespread these alleles are among the atelines and evaluating the real-world significance of the novel opsin gene mutations are essential for assessing their origin and evolutionary significance. Here, we contribute to this goal.
Our objective is (i) to clarify when and how the unique spectral property of the L/M opsins was achieved during evolution of atelines and (ii) to evaluate how effective the expanded spectral separation of the two alleles is for colour discrimination in the context of foraging ecology. For this purpose, we (i) determine nucleotide sequences and absorption spectra of L/M opsin alleles of two previously unexamined ateline species (Ateles belzebuth, Lagothrix lagotricha) and of ancestral atelines inferred, with assessing molecular structure using a protein diagram simulation and (ii) plot chromaticity of dietary fruits and background leaves in species-specific colour space based on the λmax set expected from the ‘three-sites’ rule, and the set actually observed via pigment reconstitution to quantify visual discriminability of fruits from leaves in each case.
Materials and methods
DNA samples
We sequenced the entire coding region of the L/M opsin alleles from one male and one female each of A. belzebuth and L. lagotricha. The male A. belzebuth was a captive animal housed in Primate Research Institute of Kyoto University, and his genomic DNA was extracted from a blood sample that was provided through Cooperation Research Program of the institute under their Guideline for the Care and Use of Laboratory Primates. The genomic DNA from the female A. belzebuth was extracted from a faecal sample from a wild individual in Amazonian Ecuador. The genomic DNAs of the two L. lagotricha were extracted from faecal samples provided by Yokohama Zoological Gardens, Japan. DNA extraction from the blood sample followed a conventional phenol–chloroform method. The faecal DNA was extracted by using QIAamp DNA Stool Mini Kit (Qiagen, Tokyo, Japan) as in our previous study (Hiramatsu et al. 2005). See Materials and Methods S1 (Supporting information) for PCR and DNA sequencing.
Construction of phylogenetic tree and estimation of ancestral amino acid sequences
Phylogenetic and molecular evolutionary analyses were conducted using MEGA5 (Nei & Kumar 2000; Tamura et al. 2011). The evolutionary distance (d) in terms of the number of nucleotide substitutions per site for every sequence pair was estimated using substitution models implemented in MEGA5. The L/M opsin genes of primates are known to have been homogenized between alleles by recombination or between paralogs by gene conversion, shortening their apparent evolutionary distances within a species and resulting in clustering of alleles or paralogs by species in reconstructed phylogenetic trees (Boissinot et al. 1998; Hiwatashi et al. 2011). Nonsynonymous nucleotide differences between alleles or paralogs are less susceptible to the homogenization process because of balancing selection on the alleles or purifying selection on the paralogs against gene conversion, and thus, they have been used to infer antiquity of allelic or paralogous origin of primate L/M opsin genes (Boissinot et al. 1998; Hiwatashi et al. 2011; Melin et al. 2013). We thus also evaluated the phylogenetic trees reconstructed only from nonsynonymous differences using synonymous–nonsynonymous substitution models implemented in MEGA5. The ancestral amino acid sequences at every node in the reconstructed trees were inferred by the maximum-likelihood method (Nei & Kumar 2000) implemented in MEGA5. As a mathematical model of amino acid substitution, we used the empirical substitution matrices of Dayhoff and JTT models (Dayhoff et al. 1978; Jones et al. 1992). The L/M opsin alleles of muriquis (Brachyteles) have been sequenced only partially (Talebi et al. 2006) and were therefore not included in the phylogenetic analyses.
Synthesis of woolly monkey, ancestral and mutant L/M opsin cDNAs
Site-directed mutagenesis was carried out using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, Tokyo, Japan). The cDNAs encoding the amino acid sequences of the woolly monkey L/M opsins were created by introducing mutations into the spider monkey L/M opsin cDNAs, which were previously synthesized (Hiramatsu et al. 2008). The ancestral L/M opsin cDNAs were synthesized from the woolly monkey L/M opsin cDNAs. All mutagenized cDNAs were sequenced to confirm that no spurious mutation was incorporated. See Materials and Methods S2 (Supporting information) for opsin photopigment reconstitution.
To examine the effects of amino acid substitutions on the molecular structure of the opsin, we created a three-dimensional model of the human L (red) opsin photopigment with 11-cis retinal using the structural simulation based on the ‘Swiss model’ (Stenkamp et al. 2002) [Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) (http://www.rcsb.org/pdb/home/home.do) ID: 1KPX].
Colorimetric analyses
We measured reflectance spectra for 23 fruit species (see Fig. 5 legend for species names) consumed by free-ranging spider monkeys in Santa Rosa Sector of the Área de Conservación Guanacaste, northwest Costa Rica. Although the species consumed by spider monkeys differ by geographical location, major dietary genera at Santa Rosa (such as Ficus, Brosimum, Spondias and Cecropia) are important at other sites, and our sample is a good representation of spider monkey diet that well encapsulates the range of diverse ateline foraging targets (Di Fiore et al. 2008). The chromaticity values, L/(L+M) and S/(L+M), of each specimen were given as QL/(QL+QM) and QS/(QL+QM), respectively (see Hiramatsu et al. 2008 for definition and calculation of quantum catch Qi). We calculated the chromaticity values using two sets of λmax values of the L/M opsin: one is the expected set from the ‘three-sites’ rule (560 and 550 nm) and the other is the actually observed set via photopigment reconstitution (553 and 538 nm). In both sets, the λ value of the S opsin was assumed to be 432 nm (Jacobs & Deegan 2001) as in Hiramatsu et al. 2008. We also calculated the chromaticity values of the upper and lower sides of leaves separately because those of the two sides sometimes differ dramatically (Osorio et al. 2004). Using reflectance spectra of five samples, the mean chromaticity values of fruits and upper and lower sides of leaves were calculated for each species, except for C. peltata for which only two samples were available.
Previous studies showed considerable difference of foraging efficiency to ‘conspicuous’ and ‘cryptic’ fruit categories in spider and capuchin monkeys, but the classification relied on human observation and was subjective (Hiramatsu et al. 2008; Melin et al. 2009). We objectively classified fruits into these categories relative to background leaf spectra on the basis of whether a supervised machine-learning algorism, Support Vector Machine (SVM) (Vapnik 1998), could correctly discriminate fruit from the range of leaf spectral data. The SVM was implemented using LIBSVM (Chang & Lin 2011) in MATLAB (see Melin et al. 2014 for detailed methods).
The visual discriminability of conspicuous and cryptic fruits from background leaves was then evaluated by calculating ‘just noticeable difference (JND)’ quantitative perceptual units under bright and dim light conditions. JNDs are well suited for this task because they take into consideration receptor noise and psychological limits of detection (Vorobyev & Osorio 1998). We calculated JNDs using the mean quantum catch among measured samples for each item (fruit, upper leaf and lower leaf). See Materials and Methods S3 (Supporting information) for more details of the JND calculation. We used the Wilcoxon signed-rank test (2-tailed) to compare JND values between the observed and the expected λmax sets. Statistical analyses were performed by R (ver. 2.14.1).
Results
Nucleotide sequences of the L/M opsin alleles of A. belzebuth and L. lagotricha
The L/M opsin gene sequence from the male A. belzebuth was highly similar to the SYT allele of A. geoffroyi. There was only one synonymous nucleotide difference in the entire coding region (1095 bp) (C in A. geoffroyi and T in A. belzebuth at coding nucleotide position 188). Of the two L/M opsin gene sequences possessed by the female A. belzebuth, one was identical to the SYT allele found in the male A. belzebuth and another was identical to the SFT allele of A. geoffroyi.
From the male L. lagotricha, the L/M opsin gene sequence was similar to the A. geoffroyi SYT allele (98.7% identity in the coding region; 14 nucleotide and seven amino acid differences; Fig. S1, Supporting information). The amino acid composition at sites 180, 277 and 285 of this allele remained SYT. From the female L. lagotricha, of the two L/M opsin gene sequences found, one was identical to the male sample and the other was similar to the SFT allele of A. geoffroyi (97.9% identity in the coding region; 23 nucleotide and 11 amino acid differences; Fig. S1, Supporting information). However, the three-site amino acid composition of the latter allele was AFT. The other two sites of the ‘five-sites’ rule, 197 and 308, were invariably histidine and alanine, respectively, in all the samples examined in this study.
Absorption spectra of the woolly monkey L/M opsin alleles
As the two L/M opsins of A. belzebuth were identical with those of A. geoffroyi, which were reconstituted previously (Hiramatsu et al. 2008), only the two alleles of woolly monkeys, SYT and AFT, were reconstituted in vitro. We determined their λmax to be 556 ± 1.4 nm (Fig. 1A) and 537 ± 1.4 nm (Fig. 1B), respectively. The expected λmax values of SYT and AFT L/M opsins are 560 and 545 nm, respectively (Table 1). Thus, the observed λmax values of woolly monkey SYT and AFT alleles were 4 and 8 nm shorter than expected and closer to the anomalous opsins observed previously in spider monkey SYT (553 nm) and SFT (538 nm) alleles.
Fig 1.
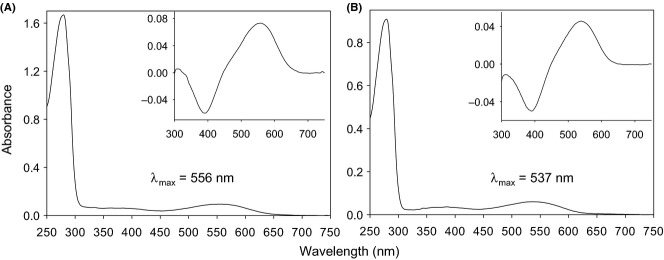
The absorption spectra of woolly monkey SYT (A) and AFT (B) types of L/M opsins measured under dark conditions. Insets: dark–light difference spectra.
Phylogenetic position of the L/M opsin alleles of spider and woolly monkeys
Using the entire coding nucleotide sequences (1095 bp), we reconstructed phylogenetic trees for the spider and the woolly monkeys and other New World monkeys representing all six types of the ‘three-sites’ composition (Table 1). The nucleotide differences were only 4% at most among the sequences. A simple neighbour-joining tree (Saitou & Nei 1987) with the Jukes–Cantor correction of evolutionary distances (Jukes & Cantor 1969) is shown as Fig. 2A. The spider monkey and woolly monkey genes formed a monophyletic clade wherein their SYT alleles formed a subclade and the SFT and AFT alleles formed another subclade. The bootstrap values in Fig. 2 were low overall, possibly because the evolutionary history differs between nucleotide sites with regards to allelic recombination (or gene conversion between paralogs in the case of howler monkeys), which homogenizes the alleles (or the paralogs) within a species. Furthermore, it is also because balancing selection on allelic difference (or purifying selection against gene conversion) may vary between nucleotide sites (Boissinot et al. 1998). Although the bootstrap supports for the ateline clade (64%) and the SYT and SFT/AFT subclades (59% and 40%, respectively) were low (Fig. 2A), the monophyletic status of the clade and the subclade pattern were not affected by application of more complex Tamura–Nei model (Tamura & Nei 1993) or the maximum-likelihood method based on the Hasegawa–Kishino–Yano model (Hasegawa et al. 1985).
Fig 2.
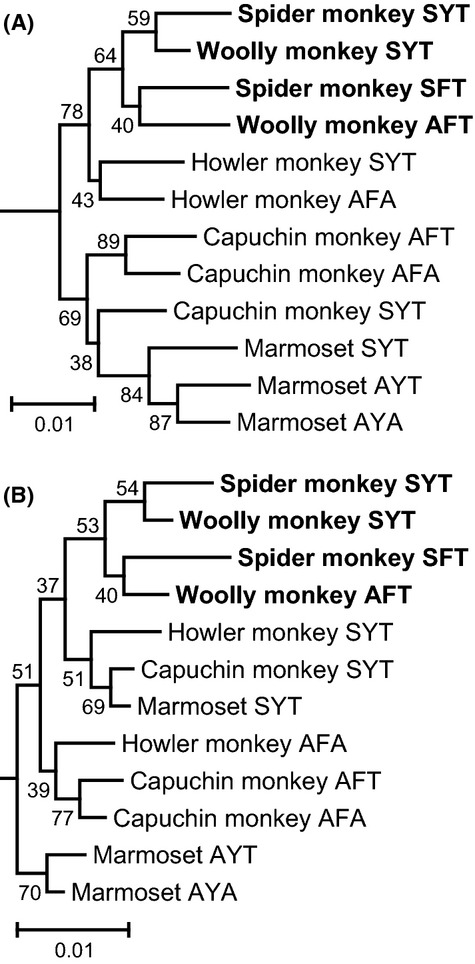
Phylogenetic trees of the L/M opsin genes reconstructed using the entire coding nucleotides (A) and nonsynonymous nucleotide differences (B) for spider and woolly monkeys and other New World monkeys representing all six types of the ‘three-sites’ composition. The phylogenetic root was given by the mouse M opsin gene (GenBank AF011389). Gap positions were removed for each sequence pair. Trees were constructed by the neighbour-joining method (Saitou & Nei 1987). The bootstrap probabilities after 1000 replication are given to each node. The species names and GenBank accession numbers: black-handed spider monkey (Ateles geoffroyi) SYT (AB193790) and SFT (AB193796); mantled howler monkeys (Alouatta palliata) SYT (AB809459) and AFA (AB809460); white-faced capuchin monkey (Cebus capucinus) SYT (AB193772), AFT (AB193778) and AFA (AB193784); and common marmoset (Callithrix jacchus) SYT (AB046546), AYT (AB046547) and AYA (AB046548). The scale bar indicates one nucleotide substitution per 100 sites.
In a phylogenetic tree (Fig. 2B) reconstructed using only nonsynonymous differences by the simple Nei–Gojobori method (Nei & Gojobori 1986), the monophyletic status of the ateline clade and the subclade pattern within it were retained. Although the bootstrap supports for the ateline clade (53%) and the SYT and SFT/AFT subclades (54% and 40%, respectively) were low (Fig. 2B), the monophyletic status of the clade and the subclade pattern were not affected by application of more complex models such as the modified Nei–Gojobori method (Zhang et al. 1998) or Li-Wu-Luo method (Li et al. 1985). In the nonsynonymous tree, genes were clustered primarily by the ‘three-sites’ types beyond species, supporting the ancient origin of allelic differentiation (Boissinot et al. 1998). The ateline clade was clustered with the SYT type of other New World monkeys, supporting that ateline SFT/AFT alleles were derived from the SYT type rather than from the AFT type seen in the capuchin monkey, which was closely related to the AFA type (Fig. 2B).
Reconstruction of ancestral opsins and spectral shifts
Based on the phylogenetic relationship between the four sequences of ateline L/M opsin alleles in Fig. 2, ancestral nodes 1–4 (A1–A4) were assigned to the tree topology (Fig. 3). We inferred the ancestral amino acid sequences at these nodes (Fig. S1, Supporting information) using the SYT type sequences of the howler monkey, capuchin monkey and marmoset as an immediate out-group cluster to the ateline clade with the topology as shown in Fig. 2B. We used the mouse M opsin sequence (Sun et al. 1997) to provide the phylogenetic root. On the basis of these ancestral sequences, amino acid substitutions were mapped on the branches of the tree (Fig. 3: for reliability of inference, see also Appendices S1–S3, Figs S2 and S3, and Table S1, Supporting information).
Fig 3.
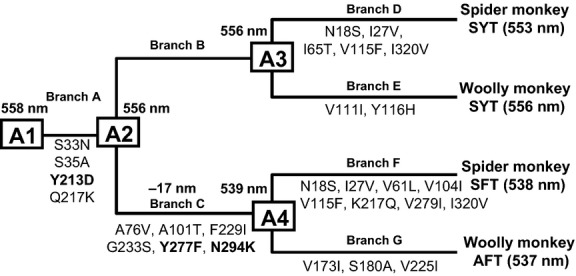
Estimation of ancestral sequences of the L/M opsin alleles of spider and woolly monkeys. The λmax values of contemporary opsins are indicated in parentheses. The λmax values of ancestral opsins are indicated at nodes A1 (Ancestor 1), A2 (Ancestor 2), A3 (Ancestor 3) and A4 (Ancestor 4). Mutations at each branch are indicated. Y213D, Y277F and N294K are highlighted with boldface letters.
The theoretical opsins A1, A2 (=A3) and A4 were reconstituted and measured for absorption spectra. The λmax of A1, A2 (A3) and A4 were 558, 556 and 539 nm, respectively (Table 2), indicating that the largest spectral shift (−17 nm) occurred at branch C, which accounts for most of the spectral difference between the current SYT and SFT/AFT alleles of spider and woolly monkeys (Fig. 3).
Table 2.
Spectral effects of mutations to opsins A1, A2 and A4
| Template | Mutants | λmax ± SD (nm) | Δλ* |
|---|---|---|---|
| A1 | 558 ± 0.8 | ||
| A1_N294K | 557 ± 1.3 | −1 | |
| A1_N294K/Y213D | 552 ± 0.8 | −6 | |
| A1_N294K/S33N/S35A | 554 ± 0.7 | −3 | |
| A1_N294K/Q217K | 555 ± 0.8 | −2 | |
| A1_N294K/S33N/S35A/Q217K | 553 ± 0.4 | −4 | |
| A1_Y213D | 552 ± 0.8 | −6 | |
| A1_S33N/S35A | 557 ± 0.9 | −1 | |
| A1_Q217K | 557 ± 1.2 | −1 | |
| A1_S33N/S35A/Y213D | 554 ± 1.3 | −4 | |
| A1_Y213D/Q217K | 552 ± 1.3 | −6 | |
| A1_S33N/S35A/Q217K | 554 ± 0.4 | −4 | |
| A1_S180A | 552 ± 1.7 | −6 | |
| A1_S180A/Y213D | 556 ± 1.0 | −2 | |
| A1_S180A/S33N/S35A | 553 ± 0.8 | −5 | |
| A1_S180A/Q217K | 553 ± 0.6 | −5 | |
| A1_S180A/S33N/S35A/Q217K | 552 ± 0.8 | −6 | |
| A1_S180A/S33N/S35A/Y213D/Q217K (A2_S180A) | 559 ± 2.0 | +1 | |
| A2 | 556 ± 2.0 | ||
| A2_Y277F | 545 ± 1.3 | −11 | |
| A2_N294K | 548 ± 0.6 | −8 | |
| A2_Y277F/N294K | 539 ± 1.2 | −17 | |
| A2_Y277F/A76V/A101T/F229I/G233S | 544 ± 0.5 | −12 | |
| A2_Y277F/F229I | 547 ± 0.2 | −9 | |
| A2_Y277F/G233S | 546 ± 0.2 | −10 | |
| A2_N294K/A76V/A101T/F229I/G233S | 547 ± 1.0 | −9 | |
| A2_Y277F/N294K/A76V/A101T/F229I | 542 ± 0.6 | −14 | |
| A2_Y277F/N294K/A76V/A101T/G233S | 541 ± 0.6 | −15 | |
| A2_Y277F/N294K/A76V/F229I/G233S | 540 ± 1.1 | −16 | |
| A2_Y277F/N294K/A101T/F229I/G233S | 541 ± 1.3 | −15 | |
| A4 | 539 ± 0.8 | ||
| A4_S180A | 538 ± 0.7 | −1 | |
| A4_V173I/V225I | 541 ± 1.2 | +2 |
Difference of λmax from the template used.
Mutations accounting for the spectral separation between SYT and SFT/AFT alleles
At branch C, there were six amino acid substitutions inferred, A76V, A101T, F229I, G233S, Y277F, and N294K (Fig. 3). Among them, the Y277F mutation caused the −11 nm spectral shift (A2_Y277F in Table 2) as expected from the ‘three-sites’ rule. Importantly, a novel mutation N294K also had a distinctive spectral effect of −8 nm (A2_N294K in Table 2). When the two mutations were introduced together, the spectral shift was −17 nm, reaching the 539 nm λmax of the opsin A4 (A2_Y277F/N294K in Table 2). When the other mutations were introduced with one or both of Y277F and N294K, the additional changes had a negligible effect (A2_Y277F/A76V/A101T/F229I/G233S–A2_Y277F/N294K/A101T/F229I/G233S in Table 2). Thus, the two mutations – Y277F and N294K – explain the evolutionary spectral shift from the opsins A2 to A4.
At branch G, there was another ‘three-sites’ mutation S180A, despite virtually no spectral shift along this branch (Fig. 3). To test whether S180A causes an expected spectral shift of approximately 5 nm and the other substitutions (V173I and V225I) have a compensatory spectral effect towards longer wavelengths, we introduced S180A and V173I/V225I separately into A4 (A4_S180A and A4_V173I/V225I, respectively). The λmax of A4_S180A was 538 nm and that of A4_V173I/V225I was 541 nm (Table 2). Thus, we verified that S180A had virtually no spectral effect at branch G.
Mutations enabling N294K spectrally effective
When N294K was introduced into the opsin A1, the spectral shift was negligible (−1 nm) (A1_N294K in Table 2). Thus, the mutation(s) at branch A enable N294K to be spectrally effective. There are four amino acid substitutions – S33N, S35A, Y213D and Q217K – inferred at branch A (Fig. 3). For convenience sake, we treated S33N and S35A, which are located closely in the N-terminal tail, as one mutation S33N/S35A. When N294K was introduced along with Y213D, the largest shift (−6 nm) occurred (A1_N294K/Y213D in Table 2). The other mutations at branch A exerted lesser spectral effects on N294K (A1_N294K/S33N/S35A–A1_N294K/S33N/S35A/Q217K in Table 2).
When Y213D was introduced to the opsin A1 without N294K (A1_Y213D in Table 2), it exerted a notable effect (−6 nm), while effects of the other mutations at branch A (A1_S33N/S35A and A1_Q217K in Table 2) were negligible. When these mutations were introduced with Y213D, there were little or no differences from Y213D alone (A1_S33N/S35A/Y213D and A1_Y213D/Q217K in Table 2). However, when these mutations were introduced together (A1_S33N/S35A/Q217K in Table 2), its spectral effect (−4 nm) was comparable to Y213D. These results indicate that mutations at branch A, Y213D in particular, are spectrally effective by themselves.
Because Y277H/N294K accounts for the spectral difference between SYT and SFT/AFT alleles, we also evaluated whether the removal of the mutations at branch A affected the λmax of A2_Y277F/N294K (Table 3). The reverse mutations N33S/A35S, K217Q and N33S/A35S/K217Q resulted in only negligible effects. Indeed, K217Q is estimated to have occurred at branch F leading to the spider monkey SFT where little spectral shift is detected (Fig. 3). The reverse mutation D213Y did not result in a measurable absorption peak in repeated experiments (A2_Y277F/N294K/D213Y in Table 3). This might suggest that this opsin is structurally unstable and that Y213D at branch A provided a safe platform for Y277F/N294K to be structurally acceptable. These results suggest that Y213D plays a major role for N294K to be effective in spectral shift of λmax.
Table 3.
Effects of mutations at branch A to opsin A2_Y277F/N294K
| Template | Mutants | λmax ± SD (nm) | Δλ |
|---|---|---|---|
| A2_Y277F/N294K | 539 ± 1.2 | ||
| A2_Y277F/N294K/N33S/A35S | 538 ± 2.5 | −1 | |
| A2_Y277F/N294K/D213Y | Failed | NA | |
| A2_Y277F/N294K/K217Q | 538 ± 2.2 | −1 | |
| A2_Y277F/N294K/N33S/A35S/K217Q | 540 ± 2.3 | +1 |
Mutations causing S180A to be spectrally ineffective
We introduced S180A into the opsin A1 and confirmed that a spectral shift towards shorter wavelengths (−6 nm) occurred (A1_S180A in Table 2), which was expected from the ‘three-sites’ rule. But when we introduced Y213D simultaneously, the spectral shift from A1 was only −2 nm (A1_S180A/Y213D in Table 2). When other mutations at branch A were introduced with S180A, their effects were lesser than Y213D (A1_S180A/S33N/S35A–A1_S180A/S33N/S35A/Q217K in Table 2). When all mutations at branch A were introduced to the opsin A1 together with S180A (A1_S180A/S33N/S35A/Y213D/Q217K in Table 2), the spectral effect of S180A disappeared (+1 nm). This mutant opsin is equivalent to A2_S180A, and the spectral shift from A2 (λmax at 556 nm) was even opposite in orientation (+3 nm). Thus, Y213D plays a major role in causing S180A to be ineffective in shifting the λmax.
Effect of Y213D and N294K in spectral shift of the human L opsin
Tyrosine at site 213 of L/M opsin is well conserved among mammals, and aspartic acid at this site has been observed in nonmammalian vertebrates and only atelines among mammals. Asparagine at site 294 is also well conserved among vertebrates, and lysine at this site has been observed in only SFT/AFT allele of spider and woolly monkeys studied here and all three alleles found in muriquis, SYT, SFT and AFA (GenBank DQ218051–DQ218055). Because of such rarity, spectral effects of these mutations on vertebrate M/LWS opsins have not been examined to our knowledge.
To investigate the generality of the spectral effects of Y213D and its effect to S180A and N294K observed in the ateline L/M opsins, we introduced these mutations into the human L (red) opsin cDNA, hs7 (Nathans et al. 1986), of which the three-site composition is SYT (Table 4). We first confirmed that the λmax of the human L opsin was 560 nm and was shifted −7 nm by S180A (Human L_S180A in Table 4) as expected from the ‘three-sites’ rule (Yokoyama & Radlwimmer 2001; Yokoyama et al. 2008). As in atelines, when Y213D was introduced together with S180A, the spectral shift became smaller (−3 nm) (Human L_S180A/Y213D in Table 4). The N294K had little spectral effect (−1 nm) (Human L_N294K in Table 4) as in ateline A1_N294K (Table 2). When Y213D was introduced together with N294K, it caused a −10 nm shift (Human L_N294K/Y213D in Table 4).
Table 4.
Spectral effects of mutations to human L opsin
| Template | Mutants | λmax ± SD (nm) | Δλ |
|---|---|---|---|
| Human L | 560 ± 0.3 | ||
| Human L_S180A | 553 ± 2.6 | −7 | |
| Human L_S180A/Y213D | 557 ± 1.2 | −3 | |
| Human L_N294K | 559 ± 5.6 | −1 | |
| Human L_N294K/Y213D | 550 ± 0.7 | −10 | |
| Human L_Y277F/N294K | 550 ± 1.0 | −10 | |
| Human L_Y277F | 553 ± 1.1 | −7 | |
| Human L_Y277F/N294K/Y213D | Failed | NA | |
| Human L_Y213D | 561 ± 2.1 | +1 |
When Y277F and N294K were introduced together, which caused −17 nm shift in the ateline A2_Y277F/N294K (Table 2), the human L_Y277F/N294K showed a −10 nm shift (Table 4). This is 3 nm larger than the observed spectral shift by Y277F alone (Human L_Y277F) (−7 nm) (Table 4) but is still within the expected range by Y277F (Yokoyama & Radlwimmer 2001; Yokoyama et al. 2008). When we introduced Y277F and N294K together with Y213D (Human L_Y277F/N294K/Y213D in Table 4), we failed to obtain a measurable absorption peak in repeated experiments. Thus, Y213D appears to be defective under Y277F/N294K in the human L opsin, while this appears to be necessary for Y277F/N294K to be structurally stable in the ateline L/M opsins (Table 3). Unlike ateline A1, Y213D itself had only minor spectral effect (+1 nm) (Human L_Y213D in Table 4). Although the effect of Y213D on S180A and N294K had some generality, its own spectral effect and its effect on other mutations (such as Y277F/N294K) could be drastically different depending on the entire amino acid sequence background.
Molecular structural modelling
Residue 213 is located near the edge of second extracellular loop (E2) nearby the fifth transmembrane domain (TM5) and residue 294 is located at the boundary between the third extracellular loop (E3) and the sixth transmembrane domain (TM6) (Fig. S1). In the three-dimensional model, we found residues 213 and 294 were closely located (Fig. 4). Electrostatic interaction of the positively charged lysine at 294 should be stronger with the negatively charged aspartic acid than with the uncharged tyrosine at 213. Thus, Y213D/N294K may cause a conformational change that could affect electrostatic environment of 11-cis retinal and λmax values of intact and mutant opsins, while N294k alone would not.
Fig 4.
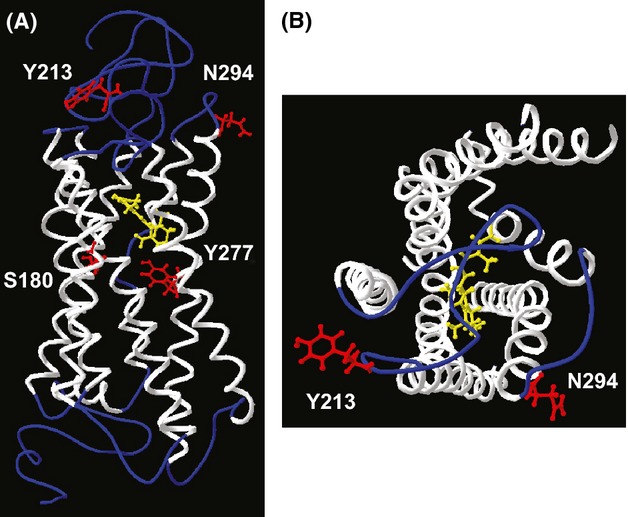
Location of residues 213 and 294 in the three-dimensional structure of the human L opsin photopigment. White helixes indicate transmembrane regions. The 11-cis retinal is depicted by yellow. The residues 180, 213, 277 and 294 are indicated by red. (A) A lateral view. N-terminal side (extracellular side) is oriented upwards. (B) Viewed from extracellular surface side.
Improvement of chromatic discrimination by mutated alleles of L/M opsins in atelines
To evaluate the effect of the expanded photopigment spectral separation in an ecological context, we compared two trichromatic phenotypes modelled with the (i) observed (553 and 538 nm) and (ii) expected (560 and 550 nm) λmax sets of the L/M opsin alleles (SYT and SFT) for visual discrimination of fruits amidst mature leaves in chromaticity plots (Fig. 5A,B). The majority of fruits were correctly classified with both the observed and the expected trichromatic models (16/23 and 17/23, respectively) (Fig. 5). We defined a fruit as a ‘conspicuous’ fruit in subsequent analyses if both models correctly discriminated it from leaves (16 species). The remaining fruits were classified as cryptic.
Fig 5.
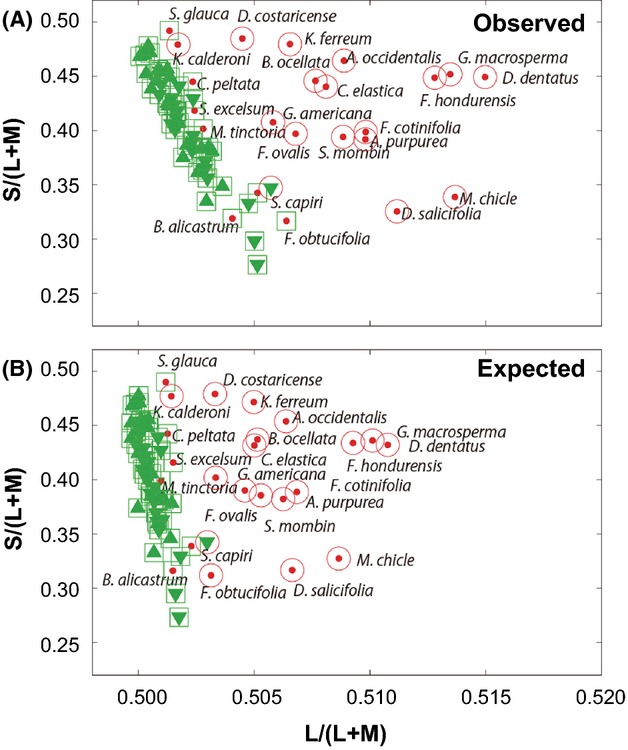
The red-green [L/(L+M)] vs. blue-yellow [S/(L+M)] chromaticity plots for fruits (red dots) and upper and lower sides of leaves (upward and downward facing green triangles, respectively) of 23 major dietary fruit species of spider monkeys under trichromat models with the observed (553 and 538 nm) (A) and the expected (560 and 550 nm) (B) λmax sets of the L/M opsin alleles (SYT and SFT). The mean values are plotted for each species. Red open circles: objects classified as fruit as by Support Vector Machine (SVM). Green open squares: objects classified as a leaf by SVM. ‘Conspicuous’ fruits in (A): Allophylus occidentalis, Annona purpurea, Bunchosia ocellata, Castilla elastic, Diospyros salicifolia, Dipterodendron costaricense, Doliocarpus dentatus, Ficus cotinifolia, F. hondurensis, F. ovalis, Genipa americana, Guettarda macrosperma, Karwinskia calderoni, Krugiodendron ferreum, Manilkara chicle and Spondias mombin. ‘Cryptic’ fruits in (A): Brosimum alicastrum, Cecropia peltata, F. obtucifolia, Maclura tinctoria, Sciadodendron excelsum, Sideroxylon capiri and Simarouba glauca. F. obtucifolia was classified as conspicuous in B and as cryptic in A.
Under the bright light condition, JND values were significantly higher for the observed trichromat model than the expected trichromat model for conspicuous fruits both against upper (Fig. 6A: P < 0.0001) and lower (Fig. 6B: P < 0.001) leaves [n = 16 pairs, Wilcoxon signed-rank 2-tailed test], but not for cryptic fruits against either leaf side [Fig. 6A, upper leaf: P = 0.22, Fig. 6B lower leaf: 0.11, n = 7 pairs]. Similar results were obtained under dim light condition (Fig. 6C,D).
Fig 6.
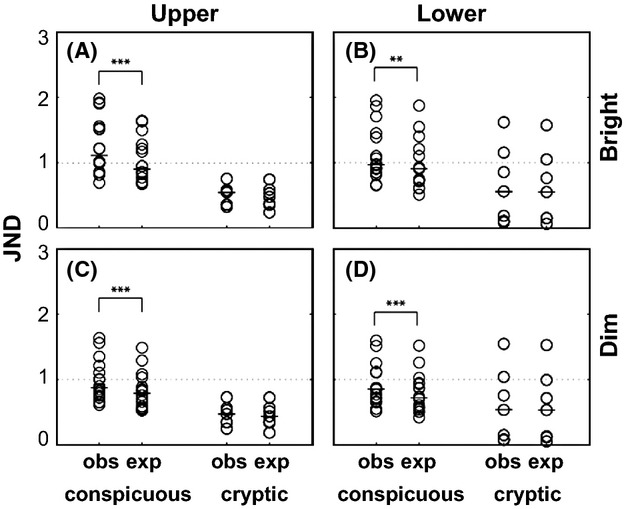
Comparison of just noticeable difference (JND) distribution between two trichromat models with the observed and the expected λmax sets of the L/M opsin alleles for conspicuous and cryptic fruits against upper leaves (A) and lower leaves (B) under bright light condition and against upper leaves (C) and lower leaves (D) under dim light condition. Each species was plotted as an open circle. Bars indicate medians. Dashed lines indicate 1 JND. Asterisks indicate significant levels, ***: P < 0.0001, **: P < 0.01, by 2-tailed Wilcoxon signed-rank test.
Theoretically, 1 JND is the smallest possible difference that can be perceived (Osorio et al. 2004; Allen & Higham 2013). Under the bright condition, the number of conspicuous fruit species over 1 JND was larger for the observed trichromat model (11 and 7 against upper and lower leaves, respectively) than the expected trichromat model (6 and 5 against upper and lower leaves, respectively) (Fig. 6A,B). Regarding cryptic fruits, there were few species over 1 JND for either trichromatic phenotype, and no difference in the number of species discriminable between the two models (0 and 2 against upper and lower leaves, respectively) (Fig. 6A,B). Similar results were obtained under the dim light condition (Fig. 6C,D).
Discussion
Here, we report novel mutations driving the evolution of vertebrate opsin genes. We show that the ‘unique’ L/M opsins previously described for black-handed spider monkeys are additionally found in a small-scale sampling from two other ateline species, common woolly monkeys and long-haired spider monkeys, implying they are widespread among ateline primates. These alleles differ from the L/M opsins of all other primates examined in that the tuning of their photopigments cannot be predicted by the ‘three-sites’ rule. We show that this departure is due to mutations that originated in the common ancestor of atelines, which changed the structural properties of the opsin and ultimately caused new amino acid residues to have a spectral effect and rendered previously important amino acids ineffective. We further showed that the enlargement of photopigment spectral separation resulting from these mutations significantly improved the discrimination of conspicuous dietary fruits from leaves in the natural habitat under both bright and dim light conditions. Both spider and woolly monkeys are specialized frugivores, with diets comprised of 55–89% fruits (see Talebi et al. 2006 and references therein). Muriquis also depend largely on fruits (21–73%) although they also depend more on young leaves (22–76%) (Talebi et al. 2006). Thus, mutations to the L/M opsins that enhance chromatic discrimination of fruits from mature leaves should be adaptive and favoured by natural selection in all three ateline genera.
Evolutionary dynamism of L/M opsin spectral types in Atelidae
Based on the present and previous findings, the current knowledge of distribution and evolutionary dynamism of L/M opsin type in the family Atelidae can be summarized as follows (Fig. 7). Atelidae consists of subfamilies Alouattinae (howler monkeys) and Atelinae. Howler monkeys have SYT and AFA opsin types as separate loci (Jacobs et al. 1996), via juxtaposition of the two alleles on the same chromosome (Boissinot et al. 1998), and are considered routinely trichromatic. Conversely, the Atelinae has polymorphic colour vision. The SYT and SFT/AFT alleles are present in all three genera, Ateles (spider monkeys), Lagothrix (woolly monkeys) and Brachyteles (muriquis) (Hiramatsu et al. 2005; Talebi et al. 2006). The AFA type is found as a rare allele in muriquis (Table 1) (Talebi et al. 2006).
Fig 7.
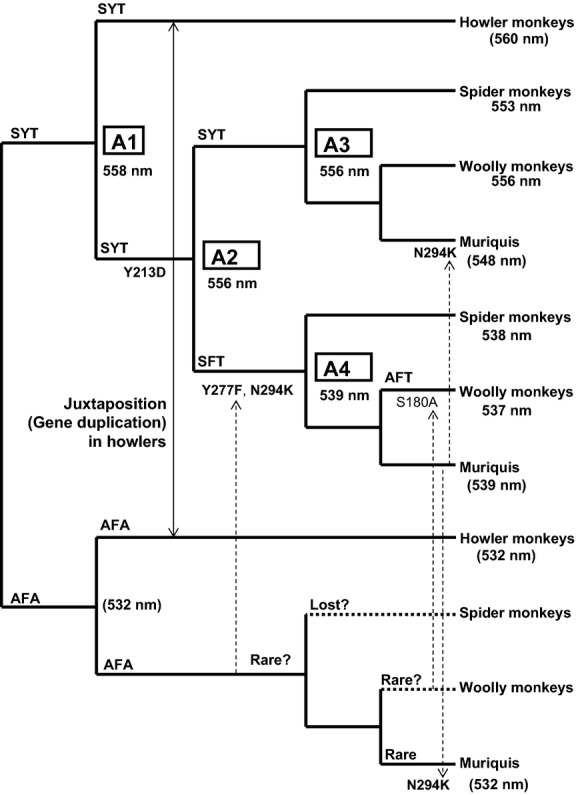
Distribution and evolutionary dynamism of L/M opsin types in the family Atelidae. The λmax values measured for contemporary and ancestral opsins are indicated following Fig. 3. The estimated λmax values are parenthesized. Values for howler monkey SYT and ancestral and contemporary AFA opsins are based on the ‘three-sites’ rule. Muriqui SYT 548 nm is based on the measured effect of N294K under Y213D. Muriqui SFT 539 nm is based on the assumption of no spectral shift from A4. Y213D, Y277F and N294K are highlighted by boldface letters. The double-headed arrow indicates juxtaposition of SYT and AFA alleles on the same chromosome in howler monkeys. Dotted arrows indicate introduction of amino acid change through recombination. Doted branches to spider monkey AFA and woolly monkey AFA indicate the possible loss or rarity of AFA allele in these genera.
The ancient origin of SYT and AFA types in New World monkeys (Fig. 2B) (Boissinot et al. 1998) and the finding of the AFA type in muriquis suggests that AFA was present in the common ancestor of the subfamily Atelinae. Derivation of the SFT allele from SYT allele (by Y277F) in the ateline common ancestor can be explained more plausibly through a regular allelic recombination with AFA allele than by a de novo point mutation. Derivation of AFT from SFT alleles (by S180A) in woolly monkeys can also be explained in the same way, with AFA allele as the donor. Thus, the AFA allele might be found as a minor allele if woolly monkey populations are extensively surveyed. Because we show that the S180A has little spectral effect in atelines, AFT may not have any selective advantage over its precursor SFT. Thus, we predict that the SFT allele should also be found in woolly monkey populations if surveyed. Sequences of muriqui SYT, SFT and AFA alleles have N294K in common (GenBank DQ218051–DQ218055). This is likely due to introduction of 294K from the SFT allele to the other two alleles through recombination. Currently, sequence information of exon 4 including the residue 213 is lacking in muriquis. Assuming that Y213D is retained in muriqui SYT, N294K could cause −8 nm shift (Table 2) and the λmax of muriqui SYT could be 548 nm (Fig. 7). Determination of entire nucleotide sequences and reconstitution of photopigments are pending for muriqui L/M opsins to further understand visual diversification in atelids.
The shortest-wave allele AFA (λmax estimated to be 532 nm; Table 1) has been found only in a northern muriqui population (Brachyteles hypoxanthus) in Brazil (Talebi et al. 2006). This allele has not been found in other muriqui populations (B. arachnoides) of southern Brazil (Talebi et al. 2006) nor in a black-handed spider monkey population in northwestern Costa Rica (Hiramatsu et al. 2005; Hiwatashi et al. 2010). Neither has evidence of the AFA allele (λmax 532 nm) been found in studies of captive spider monkeys (A. fusciceps and A. geoffroyi) and woolly monkeys (Lagothrix lagotrica) using electroretinogram (ERG) flicker photometry (Jacobs & Deegan 2001). After these robust surveys of approximately 150 X chromosomes from a diversity of species, it is reasonable to conclude that the AFA allele has been lost or is exceptionally rare in most atelines. It remains to be elucidated whether the rarity of AFA allele in atelines could be attributed to a random stochastic process.
Although more information is wanted on allele frequencies in wild populations of New World monkeys, the present study reveals previously unappreciated dynamism of colour vision variation that is exemplified in ateline New World monkeys. The finding was made possible by combination of field DNA sampling, reconstruction of contemporary and ancestral opsin photopigments and measurement of their absorption spectra, as well as colorimetric quantification of dietary fruit of monkeys in their natural habitat, emphasizing importance of multidisciplinary approach in molecular ecological studies.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research A (19207018 and 22247036) from the Japan Society for the Promotion of Science (JSPS) and Grants-in-Aid for Scientific Research on Priority Areas ‘Comparative Genomics’ (20017008) and ‘Cellular Sensor’ (21026007) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to S.K. We greatly appreciate late Prof. Osamu Takenaka for providing us with A. belzebuth genomic DNA through Cooperation Research Program of Kyoto University, and Yokohama Zoological Gardens and their staffs for L. lagotricha faecal samples. We thank Dr. James Higham and Dr. William Allen for advice on JND analysis. We thank Yusuke Hori for advices on statistical analysis.
S.K. conceived and designed research. N.O., Y.M., Y.M., R.A., M.N. and S.K. performed molecular genetic experiments. Y.M. and S.K. performed molecular evolutionary analysis. Y.M. performed molecular structural modelling. C.H. and A.D.M performed colorimetric research. C.H., A.D.M., A.D.F., C.M.S. and F.A contributed field sample collection. S.K., Y.M. and N.O. wrote the article. A.D.M and C.H. helped revise the manuscript.
Data accessibility
The nucleotide sequences of L. lagotricha SYT and AFT alleles have been deposited to the GenBank/EMBL/DDBJ databank under the accession numbers AB467314 and AB467315, respectively. The following data sets were deposited in the Dryad Digital Repository: doi: 10.5061/dryad.mg86j:1) A ‘sequential’ (fasta) format of the sequence alignment of L/M opsin genes on which all the analyses in this article is based (LMopsin_alignment.txt), 2) the dark, light and dark–light difference absorption spectra averaged among multiple measurements for each of intact and mutagenized opsins (AbsorptionSpectra.xlsx), 3) the reflectance spectra for the fruits and leaves (reflectance.xlsx), and 4) the R analysis code (JND_Wilcoxon.R) and input (JND_summary.csv) files.
Supporting Information
Additional supporting information may be found in the online version of this article.
Appendix S1 Inference of ancestral opsins.
Appendix S2 An alternative ancestral inference at site 217.
Appendix S3 An alternative scenario of ancestral sequence inference using a different out-group.
Materials and Methods S1 PCR and DNA sequencing.
Materials and Methods S2 Opsin photopigment reconstitution.
Materials and Methods S3 Calculation of JND values.
Table S1 λmax of opsins with introduced mutations under an alternative ancestral inference at site 217.
Fig S1 Amino acid sequences of the L/M opsin types found in spider and woolly monkeys and their estimated ancestral opsins.
Fig S2 An alternative ancestral inference at site 217.
Fig S3 An alternative scenario of ancestral sequence inference by different out-group.
References
- Allen WL, Higham JP. Analyzing visual signals as visual scenes. American Journal of Primatology. 2013;75:664–682. doi: 10.1002/ajp.22129. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Tan Y, Shyue SK, et al. Origins and antiquity of X-linked triallelic color vision systems in New World monkeys. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13749–13754. doi: 10.1073/pnas.95.23.13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunce JA, Isbell LA, Neitz M, et al. Characterization of opsin gene alleles affecting color vision in a wild population of titi monkeys (Callicebus brunneus) American Journal of Primatology. 2011;73:189–196. doi: 10.1002/ajp.20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine NG. Seeing red: consequences of individual differences in color vision in callitrichid primates. In: Miller LE, editor. Eat or Be Eaten. Cambridge, UK: Cambridge University Press; 2002. pp. 58–73. [Google Scholar]
- Chang CC, Lin CJ. LIBSVM: a library for support vector machines. Acm Transactions on Intelligent Systems and Technology. 2011;2:27. [Google Scholar]
- Cropp S, Boinski S, Li W-H. Allelic variation in the squirrel monkey X-linked color vision gene: biogeographical and behavioral correlates. Journal of Molecular Evolution. 2002;54:734–745. doi: 10.1007/s00239-001-0073-2. [DOI] [PubMed] [Google Scholar]
- Dayhoff MO, Schwartz RM, Orcutt BC. A model of evolutionary change in proteins. In: Dayhoff MO, editor. Atlas of Protein Sequence and Structure. Silver Spring, Maryland: National Biomedical Research Foundation; 1978. pp. 345–352. [Google Scholar]
- Deeb SS. Genetics of variation in human color vision and the retinal cone mosaic. Current Opinion in Genetics & Development. 2006;16:301–307. doi: 10.1016/j.gde.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Di Fiore A, Link A, Dew JL. Diets of wild spider monkeys. In: Campbell CJ, editor. Spider Monkeys: Behavior, Ecology and Evolution of the Genus Ateles. Cambridge, UK: Cambridge University Press; 2008. pp. 81–137. [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Hiramatsu C, Radlwimmer FB, Yokoyama S, Kawamura S. Mutagenesis and reconstitution of middle-to-long-wave-sensitive visual pigments of New World monkeys for testing the tuning effect of residues at sites 229 and 233. Vision Research. 2004;44:2225–2231. doi: 10.1016/j.visres.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Hiramatsu C, Tsutsui T, Matsumoto Y, et al. Color-vision polymorphism in wild capuchins (Cebus capucinus) and spider monkeys (Ateles geoffroyi) in Costa Rica. American Journal of Primatology. 2005;67:447–461. doi: 10.1002/ajp.20199. [DOI] [PubMed] [Google Scholar]
- Hiramatsu C, Melin AD, Aureli F, et al. Importance of achromatic contrast in short-range fruit foraging of primates. PLoS ONE. 2008;3:e3356. doi: 10.1371/journal.pone.0003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiwatashi T, Okabe Y, Tsutsui T, et al. An explicit signature of balancing selection for color-vision variation in new world monkeys. Molecular Biology and Evolution. 2010;27:453–464. doi: 10.1093/molbev/msp262. [DOI] [PubMed] [Google Scholar]
- Hiwatashi T, Mikami A, Katsumura T, et al. Gene conversion and purifying selection shape nucleotide variation in gibbon L/M opsin genes. BMC Evolutionary Biology. 2011;11:312. doi: 10.1186/1471-2148-11-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs GH. New World monkeys and color. International Journal of Primatology. 2007;28:729–759. [Google Scholar]
- Jacobs GH. Primate color vision: a comparative perspective. Visual Neuroscience. 2008;25:619–633. doi: 10.1017/S0952523808080760. [DOI] [PubMed] [Google Scholar]
- Jacobs GH, Deegan JF., II Photopigments and colour vision in New World monkeys from the family Atelidae. Proceedings of the Royal Society of London. Series B. 2001;268:695–702. doi: 10.1098/rspb.2000.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs GH, Deegan JF, II, Neitz J, Crognale MA, Neitz M. Photopigments and color vision in the nocturnal monkey, Aotus. Vision Research. 1993;33:1773–1783. doi: 10.1016/0042-6989(93)90168-v. [DOI] [PubMed] [Google Scholar]
- Jacobs GH, Neitz M, Deegan JF, Neitz J. Trichromatic colour vision in New World monkeys. Nature. 1996;382:156–158. doi: 10.1038/382156a0. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian Protein Metabolism. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- Kawamura S, Hirai M, Takenaka O, Radlwimmer FB, Yokoyama S. Genomic and spectral analyses of long to middle wavelength-sensitive visual pigments of common marmoset (Callithrix jacchus) Gene. 2001;269:45–51. doi: 10.1016/s0378-1119(01)00454-1. [DOI] [PubMed] [Google Scholar]
- Kawamura S, Takenaka N, Hiramatsu C, Hirai M, Takenaka O. Y-chromosomal red-green opsin genes of nocturnal New World monkey. FEBS Letters. 2002;530:70–72. doi: 10.1016/s0014-5793(02)03414-2. [DOI] [PubMed] [Google Scholar]
- Kawamura S, Hiramatsu C, Melin AD. Polymorphic color vision in primates: evolutionary considerations. In: Go Y, et al., editors. Post-Genome Biology of Primates. Tokyo: Springer; 2012. pp. 93–120. [Google Scholar]
- Levenson DH, Fernandez-Duque E, Evans S, Jacobs GH. Mutational changes in S-cone opsin genes common to both nocturnal and cathemeral Aotus monkeys. American Journal of Primatology. 2007;69:757–765. doi: 10.1002/ajp.20402. [DOI] [PubMed] [Google Scholar]
- Li W-H, Wu C-I, Luo CC. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Molecular Biology and Evolution. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- Melin AD, Fedigan LM, Hiramatsu C, et al. Fig foraging by dichromatic and trichromatic Cebus capucinus in a tropical dry forest. International Journal of Primatology. 2009;30:753–775. [Google Scholar]
- Melin AD, Matsushita Y, Moritz GL, Dominy NJ, Kawamura S. Inferred L/M cone opsin polymorphism of ancestral tarsiers sheds dim light on the origin of anthropoid primates. Proceedings of the Royal Society. B, Biological Sciences. 2013;280:20130189. doi: 10.1098/rspb.2013.0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin AD, Hiramatsu C, Parr NA, et al. The behavioral ecology of color vision: considering fruit conspicuity, detection distance and dietary importance. International Journal of Primatology. 2014;35:258–287. [Google Scholar]
- Mollon JD, Bowmaker JK, Jacobs GH. Variations of colour vision in a New World primate can be explained by polymorphism of retinal photopigments. Proceedings of the Royal Society of London. Series B. 1984;222:373–399. doi: 10.1098/rspb.1984.0071. [DOI] [PubMed] [Google Scholar]
- Nagao K, Takenaka N, Hirai M, Kawamura S. Coupling and decoupling of evolutionary mode between X- and Y-chromosomal red-green opsin genes in owl monkeys. Gene. 2005;352:82–91. doi: 10.1016/j.gene.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Molecular Biology and Evolution. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- Osorio D, Smith AC, Vorobyev M, Buchanan-Smith HM. Detection of fruit and the selection of primate visual pigments for color vision. The American Naturalist. 2004;164:696–708. doi: 10.1086/425332. [DOI] [PubMed] [Google Scholar]
- Saito A, Kawamura S, Mikami A, et al. Demonstration of a genotype-phenotype correlation in the polymorphic color vision of a non-callitrichine New World monkey, capuchin (Cebus apella) American Journal of Primatology. 2005;67:471–485. doi: 10.1002/ajp.20201. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shyue SK, Boissinot S, Schneider H, et al. Molecular genetics of spectral tuning in New World monkey color vision. Journal of Molecular Evolution. 1998;46:697–702. doi: 10.1007/pl00006350. [DOI] [PubMed] [Google Scholar]
- Stenkamp RE, Filipek S, Driessen CA, Teller DC, Palczewski K. Crystal structure of rhodopsin: a template for cone visual pigments and other G protein-coupled receptors. Biochimica et Biophysica Acta. 2002;1565:168–182. doi: 10.1016/s0005-2736(02)00567-9. [DOI] [PubMed] [Google Scholar]
- Sun H, Macke JP, Nathans J. Mechanisms of spectral tuning in the mouse green cone pigment. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:8860–8865. doi: 10.1073/pnas.94.16.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surridge AK, Mundy NI. Trans-specific evolution of opsin alleles and the maintenance of trichromatic colour vision in Callitrichine primates. Molecular Ecology. 2002;11:2157–2169. doi: 10.1046/j.1365-294x.2002.01597.x. [DOI] [PubMed] [Google Scholar]
- Surridge AK, Suarez SS, Buchanan-Smith HM, Smith AC, Mundy NI. Color vision pigment frequencies in wild tamarins (Saguinus spp.) American Journal of Primatology. 2005;67:463–470. doi: 10.1002/ajp.20200. [DOI] [PubMed] [Google Scholar]
- Talebi MG, Pope TR, Vogel ER, Neitz M, Dominy NJ. Polymorphism of visual pigment genes in the muriqui (Primates, Atelidae) Molecular Ecology. 2006;15:551–558. doi: 10.1111/j.1365-294X.2005.02822.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik VN. Statistical Learning Theory. Wiley: New York; 1998. [Google Scholar]
- Vorobyev M, Osorio D. Receptor noise as a determinant of colour thresholds. Proceedings of the Royal Society of London. Series B. 1998;265:351–358. doi: 10.1098/rspb.1998.0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildman DE, Jameson NM, Opazo JC, Yi SV. A fully resolved genus level phylogeny of neotropical primates (Platyrrhini) Molecular Phylogenetics and Evolution. 2009;53:694–702. doi: 10.1016/j.ympev.2009.07.019. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Molecular evolution of vertebrate visual pigments. Progress in Retinal and Eye Research. 2000;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Radlwimmer FB. The “five-sites” rule and the evolution of red and green color vision in mammals. Molecular Biology and Evolution. 1998;15:560–567. doi: 10.1093/oxfordjournals.molbev.a025956. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Radlwimmer FB. The molecular genetics of red and green color vision in mammals. Genetics. 1999;153:919–932. doi: 10.1093/genetics/153.2.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Radlwimmer FB. The molecular genetics and evolution of red and green color vision in vertebrates. Genetics. 2001;158:1697–1710. doi: 10.1093/genetics/158.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Yang H, Starmer WT. Molecular basis of spectral tuning in the red- and green-sensitive (M/LWS) pigments in vertebrates. Genetics. 2008;179:2037–2043. doi: 10.1534/genetics.108.090449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF, Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Inference of ancestral opsins.
Appendix S2 An alternative ancestral inference at site 217.
Appendix S3 An alternative scenario of ancestral sequence inference using a different out-group.
Materials and Methods S1 PCR and DNA sequencing.
Materials and Methods S2 Opsin photopigment reconstitution.
Materials and Methods S3 Calculation of JND values.
Table S1 λmax of opsins with introduced mutations under an alternative ancestral inference at site 217.
Fig S1 Amino acid sequences of the L/M opsin types found in spider and woolly monkeys and their estimated ancestral opsins.
Fig S2 An alternative ancestral inference at site 217.
Fig S3 An alternative scenario of ancestral sequence inference by different out-group.
Data Availability Statement
The nucleotide sequences of L. lagotricha SYT and AFT alleles have been deposited to the GenBank/EMBL/DDBJ databank under the accession numbers AB467314 and AB467315, respectively. The following data sets were deposited in the Dryad Digital Repository: doi: 10.5061/dryad.mg86j:1) A ‘sequential’ (fasta) format of the sequence alignment of L/M opsin genes on which all the analyses in this article is based (LMopsin_alignment.txt), 2) the dark, light and dark–light difference absorption spectra averaged among multiple measurements for each of intact and mutagenized opsins (AbsorptionSpectra.xlsx), 3) the reflectance spectra for the fruits and leaves (reflectance.xlsx), and 4) the R analysis code (JND_Wilcoxon.R) and input (JND_summary.csv) files.


