Fig 2.
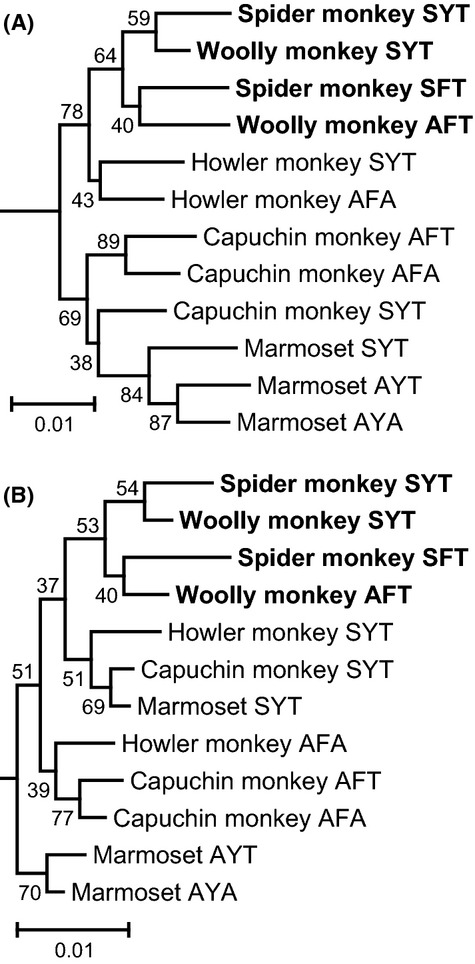
Phylogenetic trees of the L/M opsin genes reconstructed using the entire coding nucleotides (A) and nonsynonymous nucleotide differences (B) for spider and woolly monkeys and other New World monkeys representing all six types of the ‘three-sites’ composition. The phylogenetic root was given by the mouse M opsin gene (GenBank AF011389). Gap positions were removed for each sequence pair. Trees were constructed by the neighbour-joining method (Saitou & Nei 1987). The bootstrap probabilities after 1000 replication are given to each node. The species names and GenBank accession numbers: black-handed spider monkey (Ateles geoffroyi) SYT (AB193790) and SFT (AB193796); mantled howler monkeys (Alouatta palliata) SYT (AB809459) and AFA (AB809460); white-faced capuchin monkey (Cebus capucinus) SYT (AB193772), AFT (AB193778) and AFA (AB193784); and common marmoset (Callithrix jacchus) SYT (AB046546), AYT (AB046547) and AYA (AB046548). The scale bar indicates one nucleotide substitution per 100 sites.
