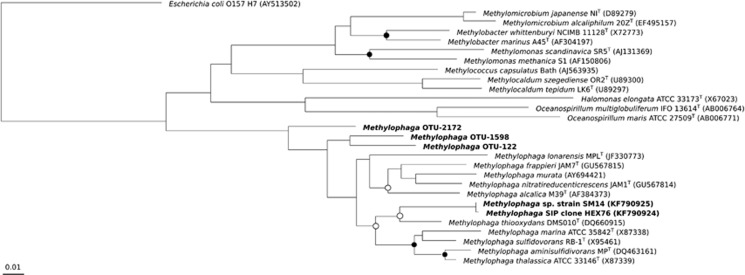
Figure 1.
Phylogenetic tree of Methylophaga operational taxonomic units (OTUs) 122, 1598 and 2172 identified in oil-contaminated plume waters at Deepwater Horizon (Rivers et al., 2013) and the hydrocarbon-degrading SIP clone HEX76 and isolated strain SM14 identified in a North Carolina surface seawater sample (Mishamandani et al., 2014). These sequences are shown in bold along with related type strains of Methylophaga and other methylotrophs in the class Gammaproteobacteria from the SILVA SSU NR 111 NR database (Quast et al., 2013). GenBank accession numbers are in parentheses. The tree was constructed using the neighbour-joining algorithm. Nodes with bootstrap support of at least 65% (○) and 90% (●) are marked (1000 replications). Escherichia coli 0157 H7 (AY513502) was used as the outgroup. The scale bar indicates the difference of number of substitutions per site.