Fig. 6.
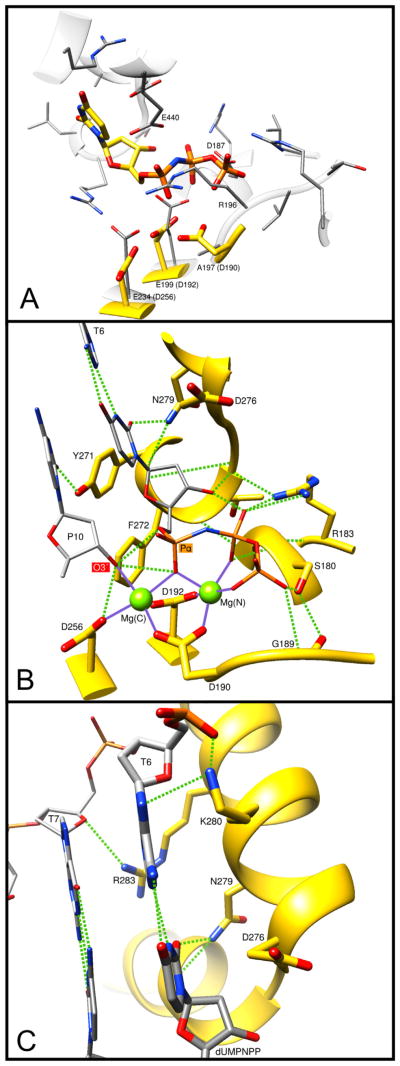
Molecular interactions of key DNA polymerase β side-chains. The ternary substrate (DNA/dUMPNPP) complex of human pol β (PDB ID 2FMS) was used to illustrate these interactions. (A) Overlay of the pol β active site (yellow carbons) with that of D. radiodurans pol X (gray carbons, PDB ID 2W9M). Key D. radiodurans residues are indicated and the equivalent pol β residues are given in parentheses. Arg196 (R196) of D. radiodurans appears to occlude the triphosphate binding pocket. In addition, pol β Asp190 (D190) is not conserved; D. radiodurans Ala197 (A197). (B) Key protein interactions with the incoming nucleotide (dUMPNPP). Hydrogen bonds are indicated with green dashed lines and metal coordinations are shown as solid purple lines. The catalytic and nucleotide binding metals are shown as green spheres: Mg(C) and Mg(N), respectively. Protein and substrate carbons are yellow and gray, respectively. The primer terminus (P10) O3′ and the α-phosphate of the incoming nucleotide are indicated. T6 identifies the templating base. The function and conservation of these residues are tabulated in Table 2 and Supplementary Table 1. (C) Key protein interactions with the bases of the nascent base pair. Hydrogen bonds are indicated with green dashed lines. T7 identifies the templating base immediately upstream of the coding templating base T6. The base of the incoming nucleotide (dUMPNPP) is indicated. The function and conservation of these residues are tabulated in Table 2 and Supplementary Table 1.