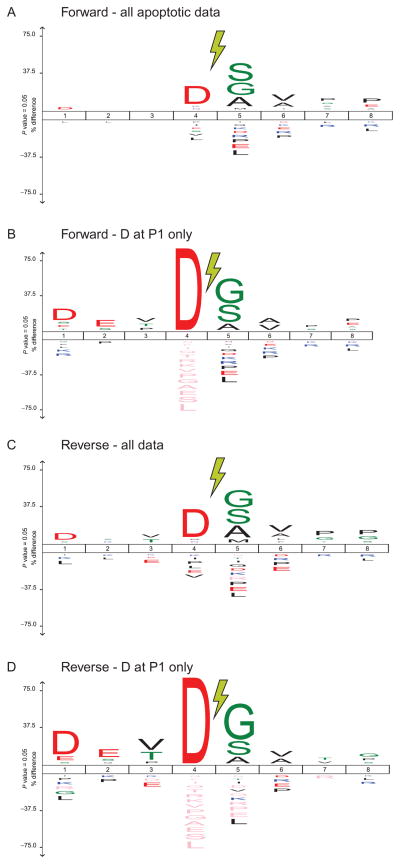
Figure 13.7.
Sequence features of identified N-termini. Aggregate plots across all N-termini shown using IceLogo, where amino acids favored at a given site are above the baseline while those disfavored are below (Colaert, Helsens, Martens, Vandekerckhove, & Gevaert, 2009). Cleavage occurs between position P1 (left of lightning bolt) and P1′. (A) Across all peptides identified in Forward Discovery experiments in apoptotic cells, Asp at P1 is highly enriched. (B) Focusing on only peptides shown in (A) with Asp at P1, a signature of caspase cleavage, we identify the canonical D-E-V-D cleavage motif for caspases from P4 to P1 site. (C and D) This motif is also conserved in N-termini identified in cell lysate incubated with recombinant caspase-3 during Reverse Discovery experiments.