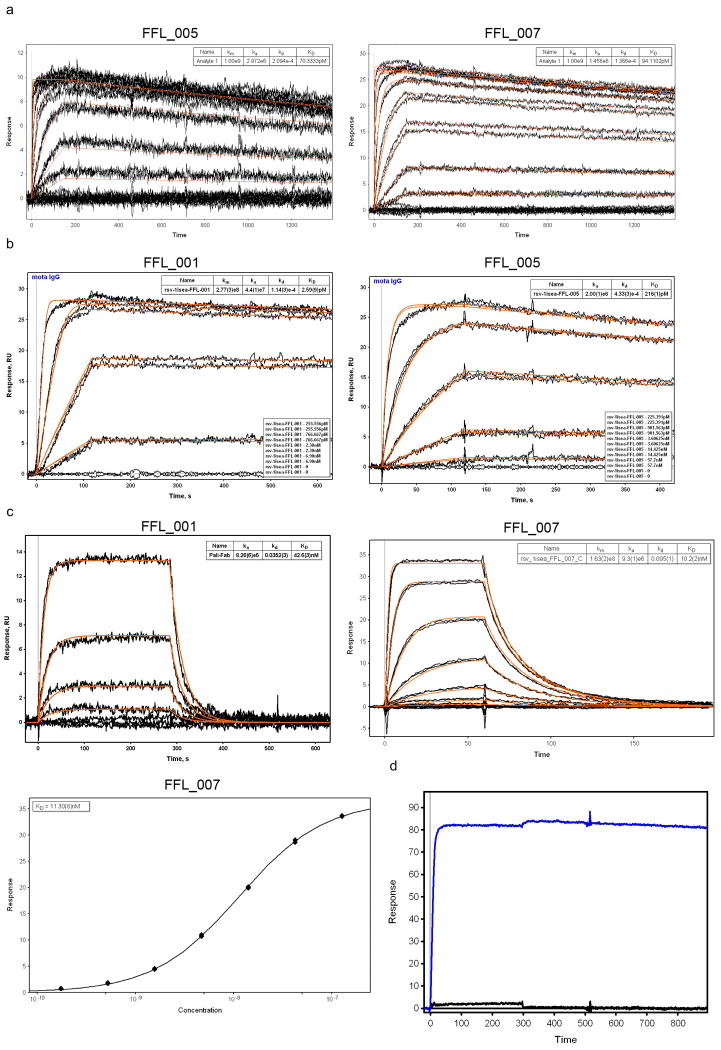
Extended Data Figure 4.
SPR data for FFL designs binding to Mota or Pali. a) Binding of FFL_005 and FFL_007 to Mota, in which the epitope scaffolds were amine-coupled to the sensor chip and Mota Fab was used as analyte. The concentrations of Mota Fab ranged from 950 nM to 436.5 pM and were used in serial dilutions with a dilution factor of three. b) Binding of FFL_001 and FFL_005 to Mota. Mota IgG was captured on the sensor chip by anti-human IgG and epitope scaffolds were used as analytes. The concentrations of scaffold ranged from 6.9 nM to 255.6 pM and were used in serial dilutions with a dilution factor of three. Kinetic fits are shown in red for both panels. c) Binding of FFL_001 and FFL_007 to Pali assessed by SPR. Pali IgG was captured by anti-human IgG on the sensor ship, and scaffolds were analytes. d) Mota binding specificity of FFL_001 assessed by SPR. Mota IgG was the ligand, captured by antihuman IgG on the sensor chip, and FFL_001 (blue) and an epitope point mutant of FFL_001 (FFL_001_K82E, black) were analytes at a concentration of 22 nM. The interaction between FFL_001 and Mota was eliminated by the point mutation.