Figure 1.
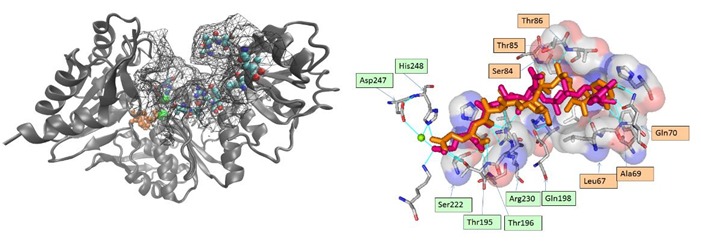
A) The M. tb Mur E enzyme (2xja) with the active site grid used for docking visible as a mesh. Critical residues of importance for natural ligand are shown as spheres. The associated ADP and magnesium in orange and green color respectively; B) Critical residues responsible for binding can be defined from the X-RAY and docked ligand: Ser222, Asp247, His248, Thr195, Thr196, Arg 230, Glu198, Leu67, Ser84, Ala69,Gln70, Thr8 and Thr85 residues interact with the natural substrate Uridine-5'- Diphosphate-N-Acetylmuramoyl-L-Alanine-D-Glutamate. LIGPLOT superimposition shows overlap at eighteen places which supports the docking algorithm being followed. The post docking ligand showed proximity with 10 out of the 13 earlier identified specificity determining residues that interact with the natural substrate. The superimposed pre and post docked conformations of the of natural substrate shows that the linear tail corresponding to the di-peptide (C21 to C28) is correctly docked in the groove of the active site (average RMSD of C21-C28 = 0.9671), while the UDP moiety has a higher RMSD of 3.753. The orange and green labels correspond to section I & II, respectively.
