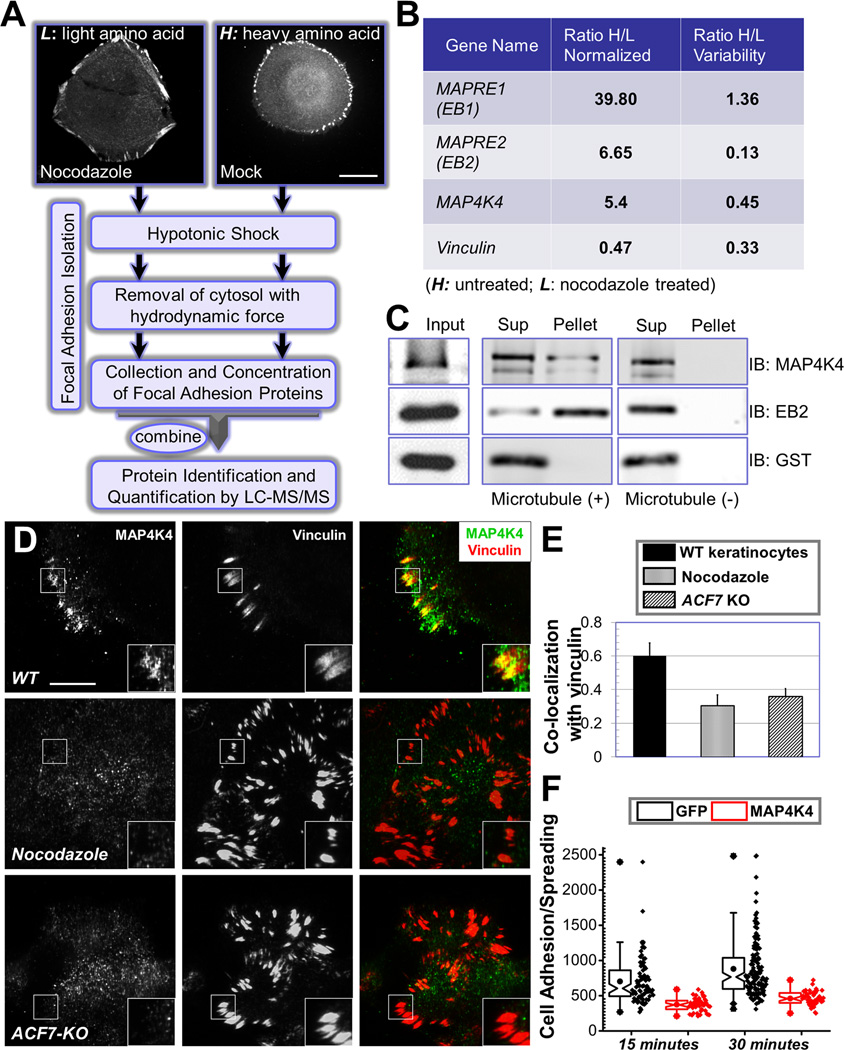
Figure 1. Identification of MAP4K4 as a MT-dependent FA protein.
(A) Flow diagram of the protocol used to isolate FA proteins for SILAC analysis. HaCaT cells were stained with α-vinculin antibody to visualize FAs (top two panels). Scale bar = 20 µm. (B) Candidates that exhibit most significant changes in protein levels upon nocodazole treatment. Ratios of each candidate protein retrieved from heavy or light cells (H/L) were shown together with the ratio variability. MAPRE1 or 2: microtubule associated protein, RP/EB family member 1 or 2. The level change of vinculin is included as a control. (C) Interaction with MTs was examined by co-sedimentation assay. Presence of protein in the original lysate (input), pellet of centrifugation as well as supernatant was determined by immunoblot with different antibodies as indicated. (D) WT mouse primary keratinocytes treated with or without nocodazole and ACF7 KO keratinocytes were fixed and subjected to immunofluorescence staining with different antibodies as indicated. Stained cells were examined by TIRF microscopy. Scale bar = 20 µm. (E) Co-localization between MAP4K4 and FAs was determined by Pearson correlation coefficient. N=10, and P<0.01. (F) GFP or GFP-MAP4K4 cells were harvested and replated onto fibronectin-coated surface for 15 or 30 minutes. Cells were then fixed and stained. Adhesion and spreading of cells (cell area) were quantified with Image J and shown as Box and Whisker plots. Box and whisker plots indicates the mean (empty square within the box), 25th percentile (bottom line of the box), median (middle line of the box), 75th percentile (top line of the box), 5th and 95th percentile (whiskers), 1st and 99th percentile (solid diamonds) and minimum and maximum measurements (solid squares), with actual data points shown at right. N>50 for each group, and P<0.01 between GFP and MAP4K4 for each time point.