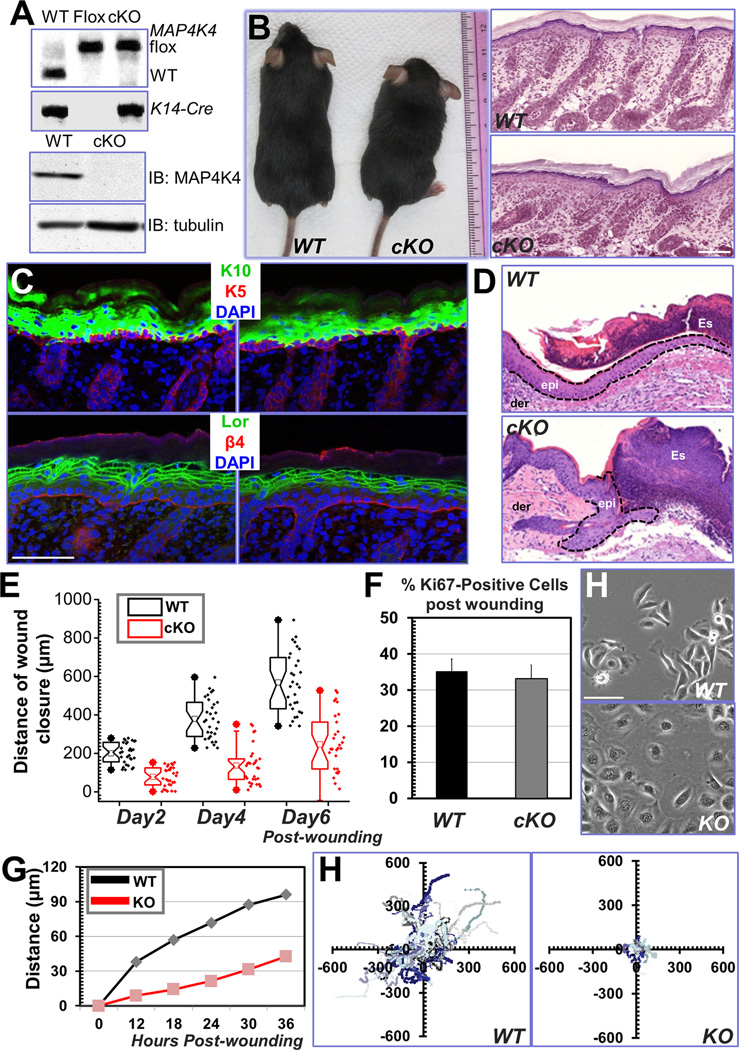
Figure 2. Cell migration defects in MAP4K4-deficient keratinocytes.
(A) PCR genotyping of MAP4K4 cKO mouse DNAs (top panel; WT: wildtype, Flox: MAP4K4fl/fl; cKO: MAP4K4fl/fl: K14-Cre). Immunoblot analysis of P0 epidermal extracts (20 µg total protein) probed with different antibodies as indicated. (B) MAP4K4 cKO mice can grow to adulthood (left panel). Histological analysis (hematoxylin/eosin staining) confirms rather normal skin and hair follicles in cKO skin (right panel). Scale bar = 50 µm. (C) Dorsal skins of neonatal mice were immunostained with different antibodies as indicated (K5: keratin 5, K10: keratin 10, Lor: Loricrin, β4: β4-integrin, CD104). Scale bar=50 µm. (D) Wound healing as monitored by histological staining of skin sections at the wound edges 4 days after injury. Halves of wound sections are shown. Epi: epidermis; der: dermis; Es: eschar. Dotted lines denote dermal–epidermal boundaries. Scale bar = 50 µm. (E) Quantification of the length of hyperproliferative epidermis generated at times indicated after wounding. Number of biological replicates (N) =30 for each group. P<0.01 for each time point. (F) Quantification of Ki67-positive cells present in wound HE. Error bars represent standard deviations (SD). N=10, P=0.22. (H) Morphology of primary keratinocytes isolated from WT or MAP4K4 cKO skin. Scale bar = 50 µm. (G) Migration of confluent monolayers of mouse keratinocytes cultured from MAP4K4 cKO and WT littermates was assessed by in vitro scratch-wound assays. The kinetics of in vitro wound healing was quantified. N=3, P<0.01. (H) Movements of individual keratinocytes were traced by videomicroscopy. Migration tracks of multiple cells for each group (WT or KO) are shown here as scatter plots. N=30.