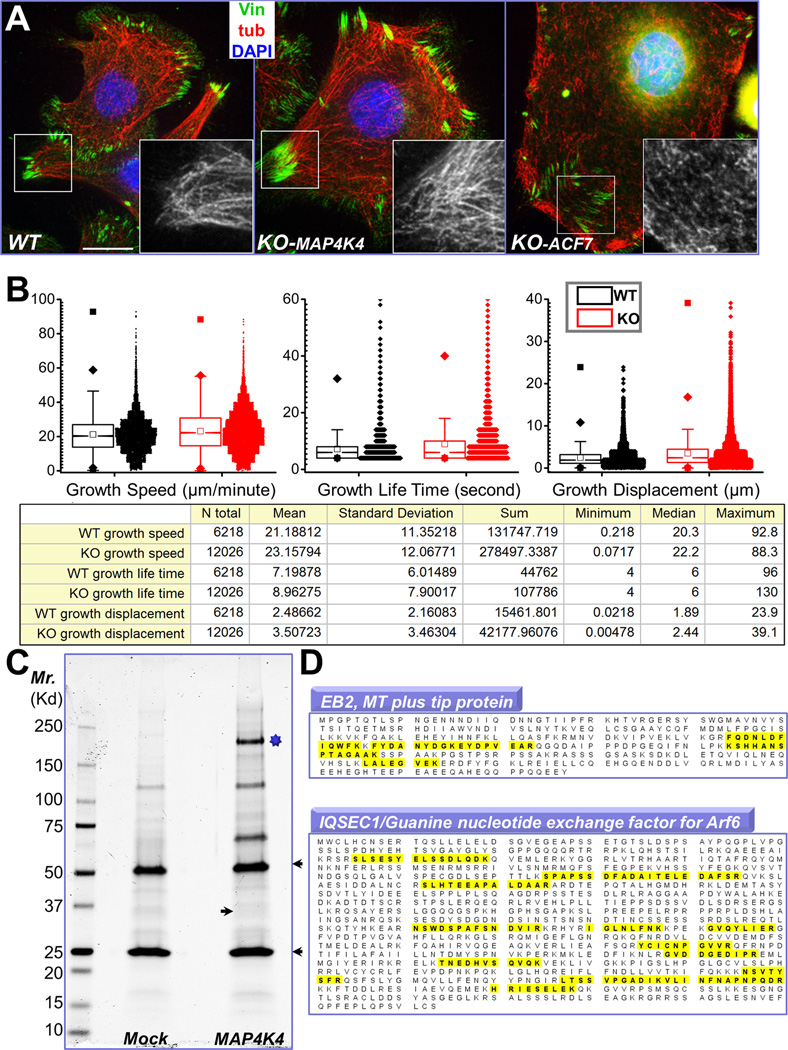
Figure 4. Identification of MAP4K4 binding partners by tandem affinity purification.
(A) Immunofluorescence for MTs (red), FAs (vinculin, green), and nuclei (DAPI, blue) shows altered MT organization in ACF7 KO cells but not in MAP4K4 deficient cells. Boxed areas are magnified as insets, where only MT staining is shown. Note bundled MT filaments merge toward peripheral FAs in WT and MAP4K4 KO cells. Scale bar = 20 µm. (B) Box and whisker plots for EB1 plus end dynamics in WT and MAP4K4 KO cells. Descriptive statistics of all the results, including mean and standard deviation, were shown in the table below. (C) MAP4K4 and its associated proteins were isolated by tandem affinity purification and resolved by SDS-PAGE. IgG heavy chain and light chain are marked by arrowheads. The putative band for MAP4K4 is marked by star. Putative band for EB2 is marked by an arrow. (D) Both EB2 and IQSEC1 were found as MAP4K4 binding proteins. Identified peptides for each protein were highlighted (yellow).