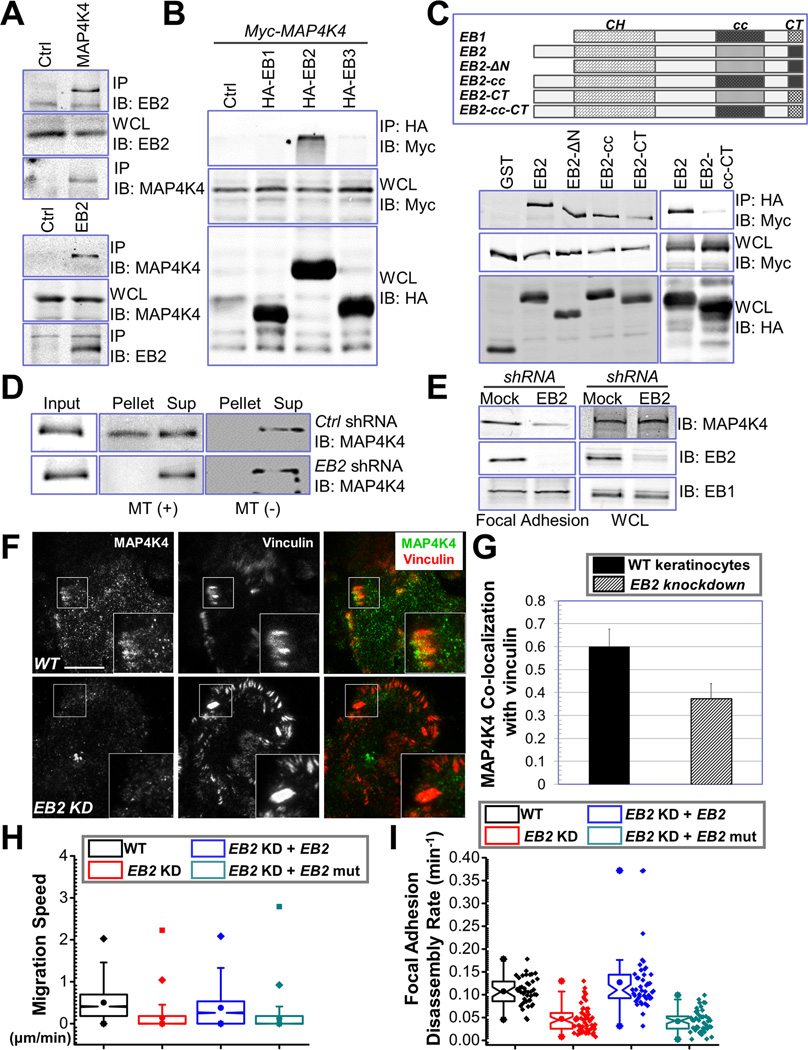
Figure 5. EB2 recruits MAP4K4 to MTs and promotes FA dynamics.
(A) Keratinocyte lysate was immunoprecipitated with α-EB2 or α-MAP4K4 antibodies. Immunoprecipitate (IP) as well as whole cell lysate (WCL, 10 µg total protein) was subjected to immunoblot with different antibodies as indicated. (B) HEK293T cells were transfected with different plasmids as indicated. Cell lysates were immunoprecipitated with α-HA antibody and immunoblotted with different antibodies as indicated. For WCL, 10 µg total proteins were used. Note only EB2 specifically pulls down MAP4K4. (C) Schematic representation of EB1, EB2, and various EB2 mutants used for coimmunoprecipitation assays (top panel). CH: calponin homology domain; cc: coiled-coil domain; CT: acidic C terminus. Association of EB2 or EB2 mutants with MAP4K4 was determined by coimmunoprecipitation as described above (lower panel). For WCL, 10 µg total proteins were used. (D) Control or EB2 knockdown cells were transfected with plasmid encoding MAP4K4. MT binding was examined by co-sedimentation assay. Pellet and supernatant as well as an aliquot of pre-cleared cell lysate (input) were immunoblotted to assess the level of MAP4K4. (E) Presence of MAP4K4, EB2, and EB1 in isolated FAs or whole cell lysate (WCL, 20 µg) from control or EB2-shRNA treated cells was determined by immunoblots. (F–G) WT keratinocytes and EB2 knockdown (KD) cells were subjected to immunofluorescence staining and examined by TIRF microscopy (F). Co-localization between MAP4K4 and vinculin was determined by Pearson correlation coefficient and quantified (G). Scale bar = 20 µm. N=10, P<0.01. (H–I) Quantifications of migration velocities and FA disassembly for control, EB2-knockdown (KD) cells, and cells rescued with WT EB2 or EB2 mutant (EB2-cc-CT). Note that only WT EB2 restored FA dynamics and rescued the speed of migration. For cell motility assay (I), N= 30 cells × 120 time points for each group. P<0.01 between WT and EB2 KD; EB2 KD and EB2 KD + EB2; and EB2 KD + EB2 and EB2 KD + EB2 mutant. For FA disassembly (J), N>40 for each group. P<0.01 between WT and EB2 KD; EB2 KD and EB2 KD + EB2; and EB2 KD + EB2 and EB2 KD + EB2 mutant.