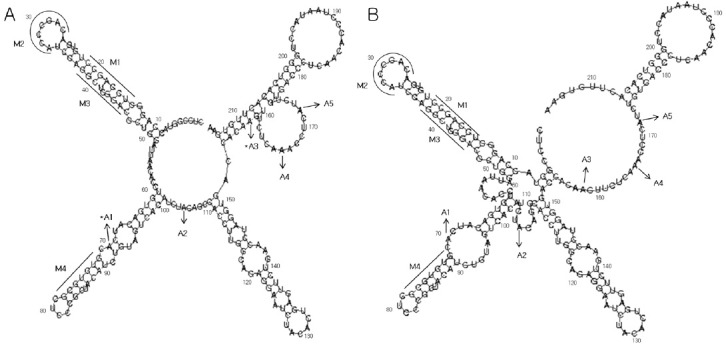
Fig. 3. Most single-stranded regions matched the computer modeling predictions. The secondary structure of Irf7 5’-UTR1- 215 was predicted using Vienna fold, which calculates both minimal free energy and base-pairing possibilities. (A) The plain sequence of the Irf7 5’UTR was applied with no constraint. The experimentally identified single-stranded regions are indicated by a series of acylations (A). The A2, A4, and A5 regions were identified as unpaired regions, confirming the computer predictions; however, regions A1 and A3 did not match the computer predictions. The asterisk indicates the mismatched areas. Mutated regions are indicated with an ‘M’. In M1 and M4, the double-stranded sequences UCCAGCC and GUGUGCG were mutated to single-stranded AGGUCGG and CACACGC, respectively. In M2, the unpaired CAGCCCA was changed to the complementary sequence. M1 and M3 were converted to the complementary sequence but the double-stranded structure was retained. (B) The experimentally identified single-stranded regions were constrained.