Figure 1.
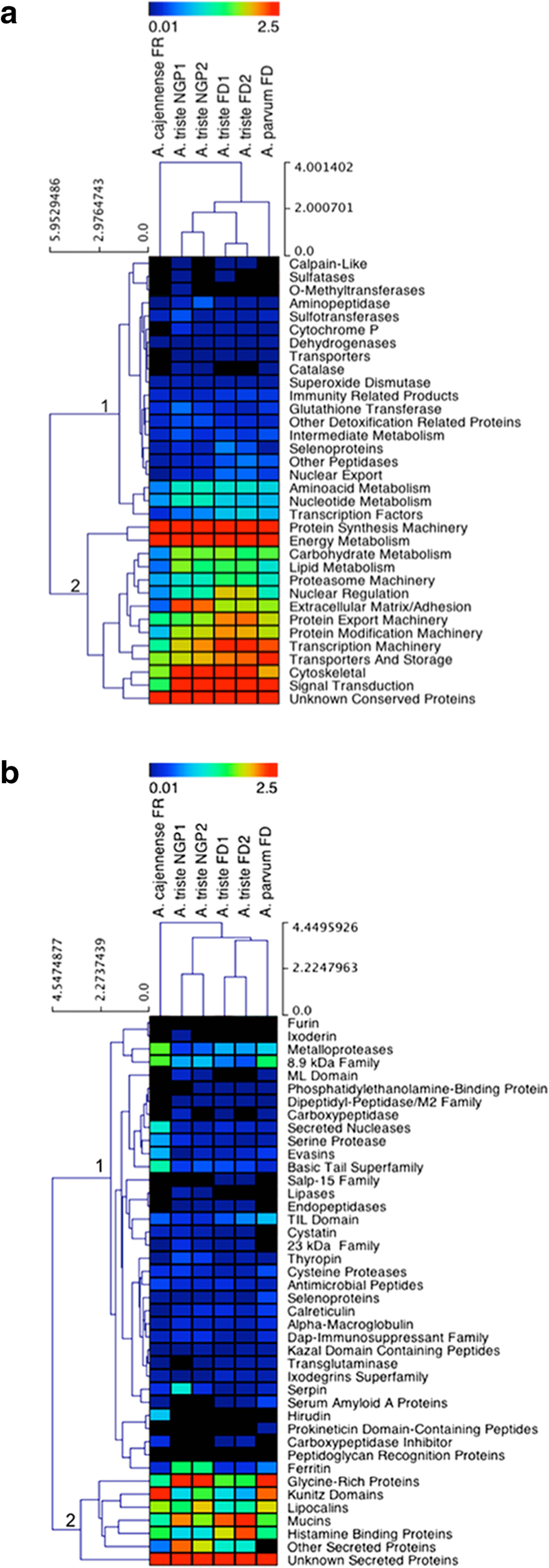
Hierarchical clustering of putative protein families from the sialotranscriptome of Amblyomma ticks. The heatmaps display the expression level of the protein families (rows) for each sialotranscriptome (columns). a) Families classified as housekeeping proteins. b) Families classified as secreted proteins. The color scale represents the log2 transformation of % of reads + 1. The transition of colors from blue to red represents an increase in the % of reads. Black color indicates the % of reads <0.01; red color indicates the % of reads ≥ 4.5. The length of branches in hierarchical trees indicates the degree of similarity between objects, either tick libraries (columns) or protein families (rows). Side scales represent the node height (cluster-to-cluster distance). NGP1: nymphs fed on tick-naïve guinea pigs; NGP2: nymphs fed on guinea pigs undergoing a second infestation; FD1: females fed on tick-naïve dogs (first infestation) and FD2: females fed on dogs undergoing a second infestation. FR: females fed on rabbits. The clustering method used Euclidian distance with average linkage.
