Figure 4.
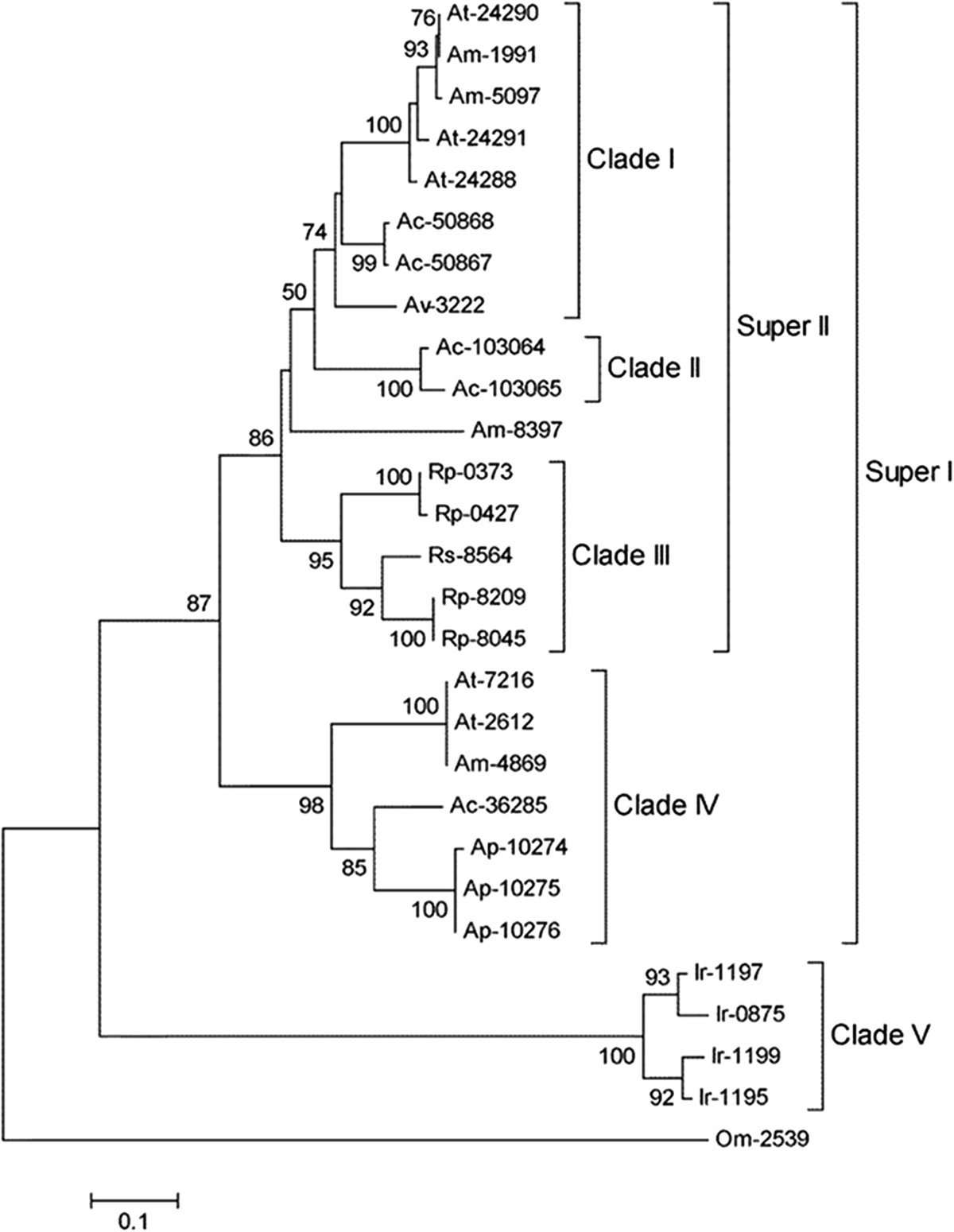
Phylogenetic analysis of the salivary thyropin proteins from different tick species. Phylogram resulting from the alignment of 11 full-length protein sequences from the sialotranscriptomes of A. triste (At-7216, At-2612, At-24288, At-24290 and At-24291), A. parvum (Ap-10275, Ap-10276 and Ap-10274) and A. cajennense (Ac-103065, Ac-50867 and Ac-36285) as well as sequences from other ticks previously deposited in the NCBI protein database, including A. maculatum (Am-1991, Am-8397, Am-5097 and Am-4869), A. variegatum (Av-3222), Ixodes ricinus (Ir-1197, Ir-0875, Ir-1199 and Ir-1195), Rhipicephalus pulchellu s (Rp-0373, Rp-8209, Rp-0427 and Rp-8045), R. sanguineus (Rs-8564) and Ornithodoros moubata (Om-2539). The database access numbers used in this analysis are listed in Additional file 5: AF5. The neighbor-joining method with 1,000 bootstrap replicates was used for analysis in MEGA 5.05 software. The numbers on the nodes indicate the bootstrap support. Brackets highlight branches with more than 72% bootstrap support. The bar at bottom indicates 10% amino acid divergence.
