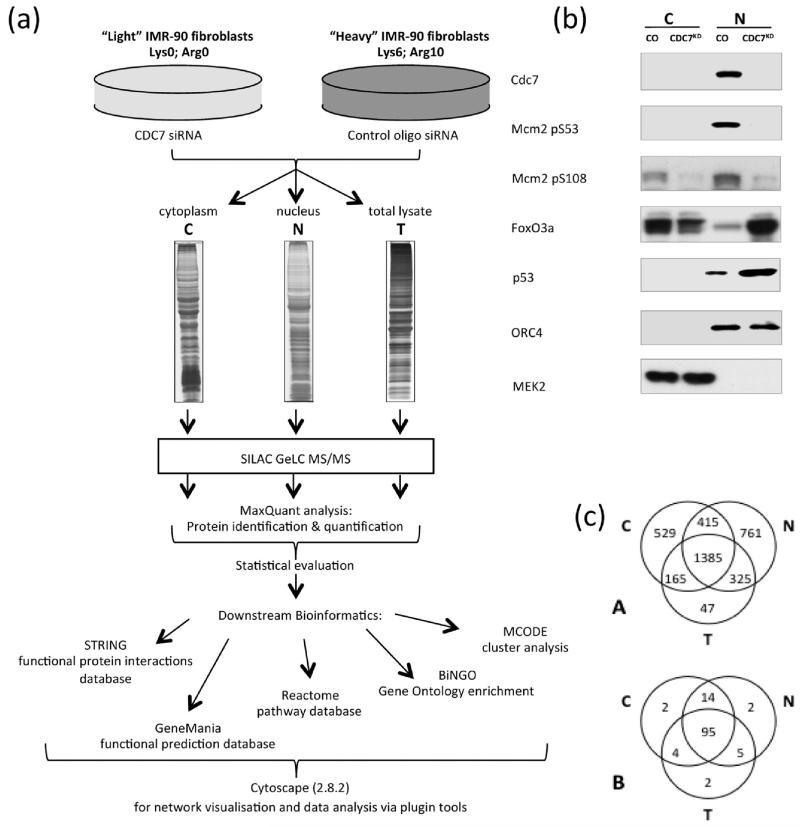
Figure 1.
Comparative analysis of CDC7-depleted nuclear-enriched and cytoplasmic-enriched fractions. (a) SILAC workflow: SILAC-labeled IMR90 fibroblasts were treated with CDC7 siRNA or control oligo and fractionated into cytoplasmic, nuclear or total cell lysates. MaxQuant software was used for protein inference and quantitation.24 STRING,26 GeneMania30 and Reactome32 software were used for protein networks, functional annotation enrichment and pathway analysis. Cytoscape28 was used for network visualization and for further analysis with BiNGO34 and MCODE33 plugins. (b) Cells treated with either control oligo or CDC7-siRNA were fractionated into cytoplasmic and nuclear-enriched fractions and subjected to Western blot analysis for CDC7, phospho-MCM2 (pS53 and pS108 sites), p53, FOXO3a, ORC4 and MEK2. (c) Venn diagram showing: (A) distribution over the three sample types for the 3627 proteins identified; (B) distribution over the three sample types for the strict set of 124 significantly changed proteins (see text).