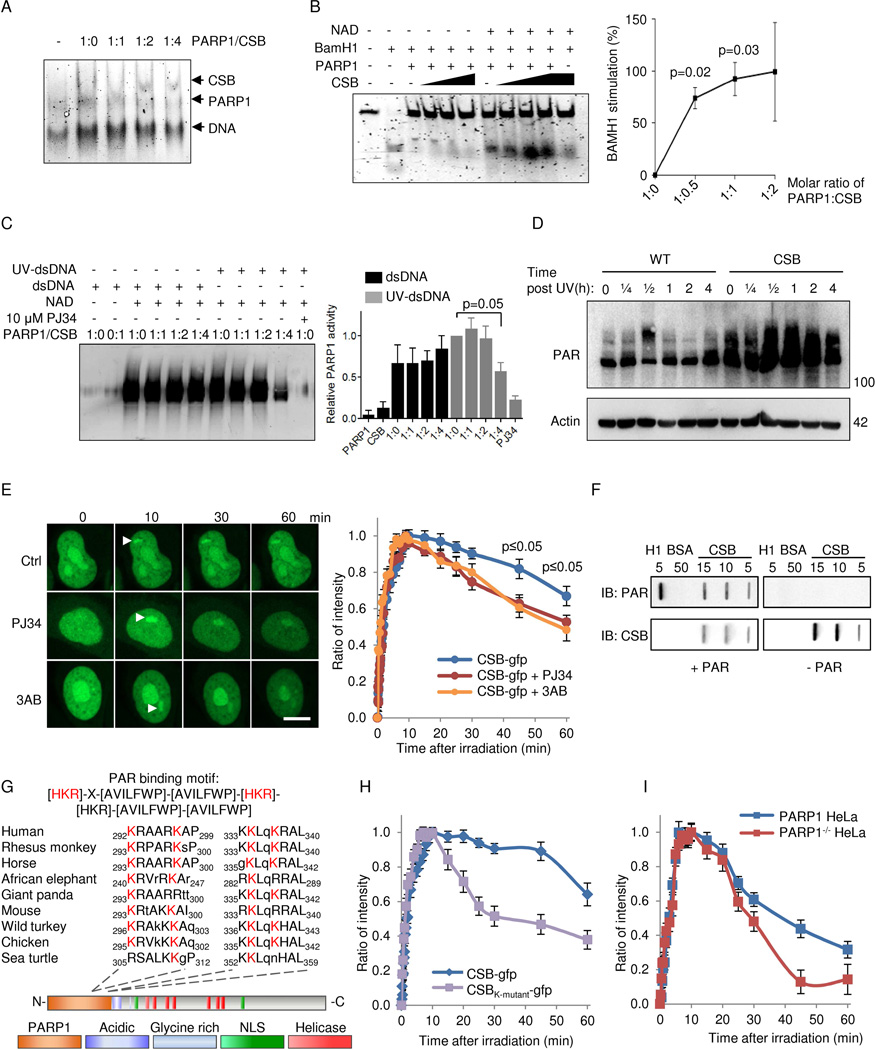
Fig. 6. CSB inhibits PARP1 activation in vitro and in vivo through displacement of PARylated PARP1 from DNA.
(A) Electromobility shift assay (EMSA) of PARP1 and CSB binding to double stranded DNA. (B) BamH1 incision of a 42-mer oligo preincubated with 250 nM PARP1 and with increasing amounts of CSB with or without NAD+ (n=3, mean ± SEM). (C) Representative immunoblot and quantification of in vitro poly-ADP-ribosylation (PARylation) of CSB and PARP1. Reactions were performed with recombinant proteins in the presence of undamaged or damaged DNA (n=3, mean ± SEM). (D) Representative immunoblot of whole cell (PARylation) after 5 J/m2 treatment in WT and CSB deficient cells at various timepoints. (E) Recruitment of gfp-tagged CSB to laser induced DNA damage after 1 hour preincubation with PARP inhibitors 3AB or PJ34. (F) Representative slot-blot showing non-covalent interaction between CSB and PAR. (G) The PAR binding motif in CSB. (H) Recruitment to laser induced DNA damage of gfp-tagged WT CSB or CSB harboring K to A mutations in the 4 conserved lysines. (I) Recruitment of gfp-tagged WT CSB to laser induced DNA damage in WT HeLa or PARP1−/− HeLa cells.