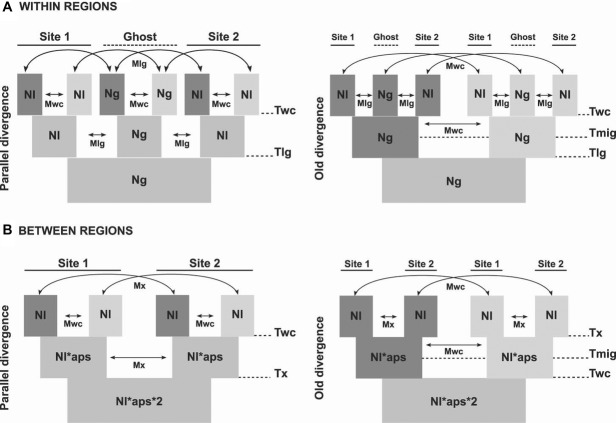
Figure 3.
Historical demographic models compared using ABC. (A) Two sites within the same region. (B) Two sites, each in a different region. Dark shading indicates Crab ecotype, light shading the Wave ecotype and intermediate shading the ancestral populations. 1, 2—sampled localities; arrows signify migration. The present is represented at the top of each diagram.Parameters used were: Within-region models: Ng – effective size of the ancestral and ghost population, Nl – effective size of a local population, Tlg – time of separation of the spatially separated populations, Twc – time of separation of the ecotype populations, Tmig (old divergence model only) – time since the end of allopatric separation of ecotypes, PROPT – (parallel model only) – log(Twc)/log(Tlg), PROPTlg (old divergence model only) – log(Tlg)/log(Twc), PROPmig (old divergence model only) – log(Tmig)/log(Twc), Mlg – probability of an individual migrating between a local and a ghost population (no direct migration allowed between local populations), Mwc – probability of an individual migrating between populations of the different ecotypes. For the old divergence model, the constraint PROPmig>PROPTlg was imposed. Between-region models: Nl – effective size of a local population, APS – relative size of the ancestral population, Tx – time of separation of the regional populations, Twc – time of separation of the ecotype populations, Tmig (old divergence model only) – time since the end of allopatric separation of ecotypes, PROPT (parallel model only) – log(Twc)/log(Tx), PROPTx (old divergence model only) – log(Tx)/log(Twc), PROPmig (old divergence model only) – log(Tmig)/log(Twc), Mx – probability of an individual migrating between populations in different regions, Mwc – probability of an individual migrating between populations of the different ecotypes. For the old divergence model, the constraint PROPmig>PROPTx was imposed.