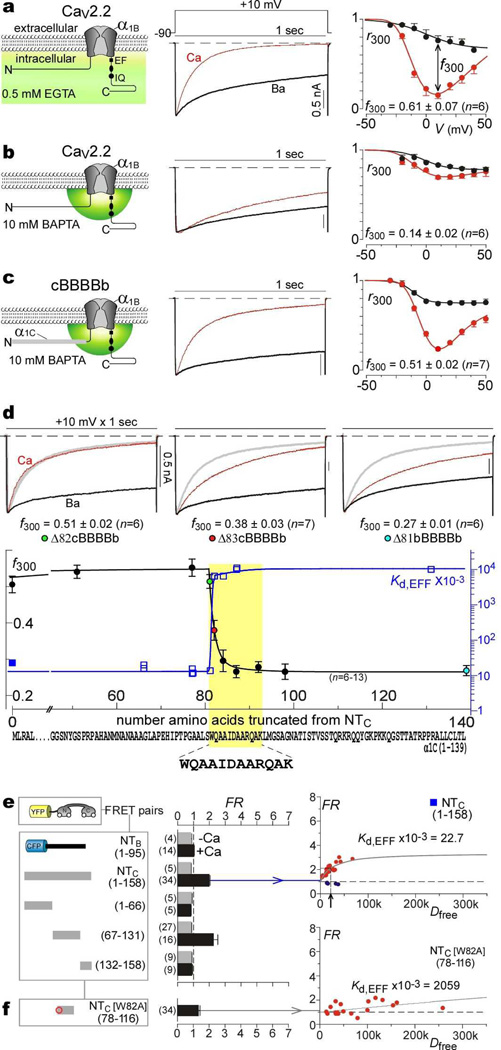
Figure 1. Transformation of spatial Ca2+ selectivity in CaV2.2 channels.
a, CaV2.2 CDI, low buffering. Left, cartoon of global Ca2+ elevation. Middle, exemplar traces in Ca2+ (red) and Ba2+ (black). Throughout, current bar references Ca2+ trace; Ba2+ trace normalized to Ca2+ peak. Right, average CDI; f300 at 10 mV; data, mean ± SEM throughout; cell number in parentheses. b, CaV2.2 CDI, high buffering. Left, cartoon of local Ca2+ signaling. c, CDI of cBBBBb. d, Localizing spatial Ca2+ transforming element (yellow highlight) within NTC. Top, exemplar currents for cBBBBb with amino-terminal deletions, (Δ82cBBBBb, removal of first 81 aa of cBBBBb; Δ81bBBBBb, removal of entire amino terminus of CaV2.2). Bottom, f300 (high buffering) versus residues deleted from cBBBBb; left axis and circles with colors corresponding to exemplars above. Kd,EFF is also plotted on same abscissa; blue right axis and blue squares, with filled symbol corresponding to e. Extreme bottom, localized sequence for both CDI transformation and Ca2+/CaM binding within NTC (yellow highlight).
e, f, FRET for YFP–CaM versus channel amino-terminal segments tagged with CFP. Left, construct schematics. Middle, FRET ratios (FR). Right, exemplar binding curves, with red symbols for Ca2+/CaM, and blue symbols for apoCaM (e, arrow shows Kd,EFF).