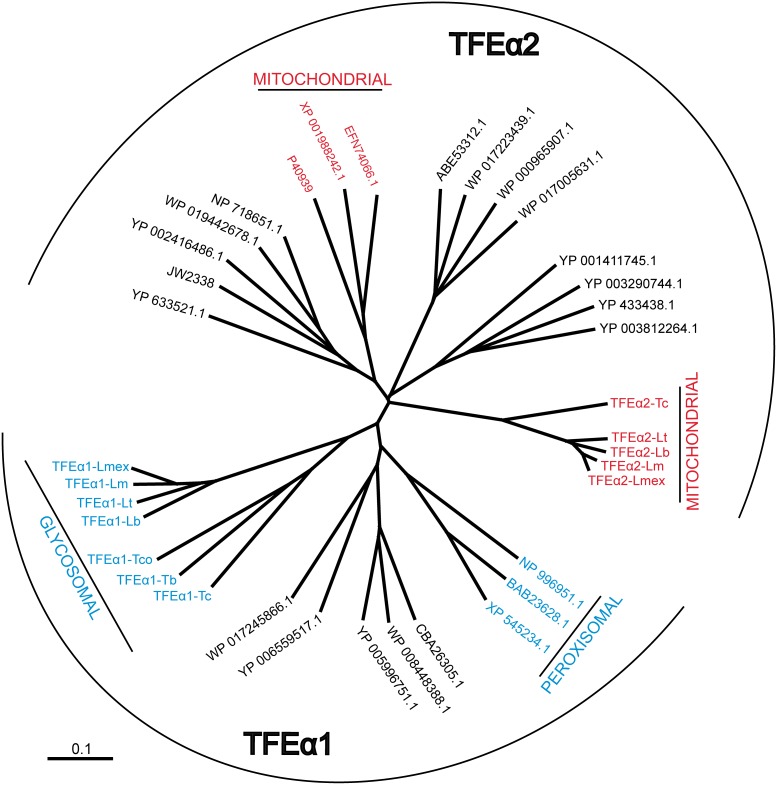
Figure 4. Dendrogram of trifunctional enzyme (TFE) isoforms.
Prokaryotic (black characters) and eukaryotic (colored characters) TFEα sequences are represented by their GenBank accession codes. Glycosomal/peroxisomal (TFEα1) or mitochondrial (TFEα2) proteins are highlighted in blue and red. Experimental evidence for glycosomal localization of trypanosomatid TFEα1 isoforms, which all contain a PTS2 motif, is limited to T. brucei TFEα1 (see [40] and S4 Figure). Mitochondrial localization of the trypanosomatid TFEα2 isoforms is assumed due to an N-terminal mitochondrial targeting motif and the absence of a PTS motif. Abbreviations: Lb, Leishmania braziliensis; Lm, L. major; Lmex, L. mexicana; Lt, L. tarentolae; Tb, T. brucei; Tc, T. cruzi; Tco, T. congolense. The organisms corresponding to the accession numbers are: Canis lupus familiaris (XP_545234.1), Danio rerio (NP_996951.1), Mus musculus (BAB23628.1), Curvibacter putative symbiont of Hydra magnipapillata (CBA26305.1), Janthinobacterium sp. HH01 (WP_008448388.1), Marinobacter sp. BSs20148 (YP_006559517.1), Pseudomonas stutzeri (WP_017245866.1), Ralstonia solanacearum CMR15 (YP_005996751.1), Camponotus floridanus (EFN74066.1), Drosophila grimshawi (XP_001988242.1), γ-proteobacterium HdN1 (YP_003812264.1), Hahella chejuensis KCTC2396 (YP_433438.1), Homo sapiens (P40939), Moritella dasanensis (WP_017223439.1), Parvibaculum lavamentivorans DS-1 (YP_001411745.1), Rhodothermus marinus DSM4252 (YP_003290744.1), Shewanella denitrificans OS217 (ABE53312.1), Vibrio splendidus LGP32 (YP_002416486.1), Escherichia coli (JW2338), Enterovibrio norvegicus (WP_017005631.1), Moritella marina (WP_019442678.1), Myxococcus xanthus DK1622 (YP_633521.1), Shigella flexneri (WP_000965907.1), Shewanella oneidensis MR-1 (NP_718651.1).