Abstract
Sortase, a cysteine-transpeptidase conserved in Gram-positive bacteria, anchors on the cell wall many surface proteins that facilitate bacterial pathogenesis and fitness. Genetic disruption of the housekeeping sortase in several Gram-positive pathogens reported thus far attenuates virulence, but not bacterial growth. Paradoxically, we discovered that depletion of the housekeeping sortase SrtA was lethal for Actinomyces oris; yet, all of its predicted cell wall-anchored protein substrates (AcaA-N) were individually dispensable for cell viability. Using Tn5-transposon mutagenesis to identify factors that upend lethality of srtA deletion, we uncovered a set of genetic suppressors harboring transposon insertions within genes of a locus encoding AcaC and a LytR-CpsA-Psr (LCP)-like protein. AcaC was shown to be highly glycosylated and dependent on LCP for its glycosylation. Upon SrtA depletion, the glycosylated form of AcaC, hereby renamed GspA, was accumulated in the membrane. Overexpression of GspA in a mutant lacking gspA and srtA was lethal; conversely, cells overexpressing a GspA mutant missing a membrane-localization domain were viable. The results reveal a unique glycosylation pathway in A. oris that is coupled to cell wall anchoring catalyzed by sortase SrtA. Significantly, this novel phenomenon of glyco-stress provides convenient cell-based assays for developing a new class of inhibitors against Gram-positive pathogens.
INTRODUCTION
Gram-positive pathogens utilize a cysteine-transpeptidase enzyme known as sortase to covalently attach virulence factors to peptidoglycan for surface display (Ton-That et al., 2004). The first sortase termed SrtA was discovered in Staphylococcus aureus (Mazmanian et al., 1999). Substrates of this enzyme harbor an N-terminal signal peptide and a C-terminal cell wall sorting signal (CWS), which is comprised of a LPXTG motif, followed by a stretch of hydrophobic and positively charged residues (Mazmanian et al., 2001). The cell wall anchoring of a surface protein begins by the synthesis of the protein precursor in the cytoplasm, which is translocated across the membrane by the Sec machinery. The transported protein is laterally inserted into the membrane via its hydrophobic domain for recognition and capture by sortase. Sortase catalyzes a transpeptidation reaction by cleaving the LPXTG motif between the threonine and glycine residues and linking the terminal threonine moiety of the cleavage product to the stem peptide of a Lipid II precursor molecule. The incorporation of this cell wall precursor into a growing chain of peptidoglycan allows surface display of the sortase substrates (Ton-That et al., 2004).
S. aureus SrtA belongs to the class A family of sortase enzymes (Comfort & Clubb, 2004, Dramsi et al., 2005) that utilize the hydrophobic amino terminus with the signal sequence as the transmembrane domain and anchor surface proteins to the cell wall as monomers by the mechanism described above. Many Gram-positive pathogens, such as Corynebacterium diphtheriae, Enterococcus faecalis, Bacillus cereus, and streptococci, produce another class of sortase enzymes, referred to as class C sortases (Ton-That & Schneewind, 2003), which are inserted in the membrane via an extended carboxy terminal domain having a trans-membrane motif. These sortases are specific for the assembly of protein polymers called pili or fimbriae (Mandlik et al., 2008). Importantly, the pilus polymers are ultimately anchored to the cell wall by the nonpolymerizing, class A sortase or the housekeeping sortase (Swaminathan et al., 2007, Budzik et al., 2007, Nobbs et al., 2008, Sillanpaa et al., 2013, Nielsen et al., 2013, Smith et al., 2010). Because many surface proteins and pili are cell wall anchored virulence determinants, genetic disruption of class A sortase genes is expected to attenuate bacterial virulence (Mazmanian et al., 2000, Bierne et al., 2002, Chen et al., 2005, Bolken et al., 2001). Initially, the S. aureus sortase gene srtA was identified from a screen of temperature-sensitive mutants with anticipation that it would be an essential gene (Mazmanian et al., 1999, Novick, 2000). However, none of the sortase mutants including deletions that have been reported thus far has any major growth defects or aberrant cell morphology. This is somewhat surprising as class A sortases are considered to carry out a “housekeeping” function (Scott & Zahner, 2006) and their substrates are variously associated with membrane translocation and cell wall synthesis pathways (Ton-That & Schneewind, 1999, Perry et al., 2002, Ruzin et al., 2002).
This paradox is further highlighted in the current study with Actinomyces oris, a key Gram-positive colonizer of the oral cavity, which helps to seed dental plaque. Our prior BLAST search for sortase homologs in the genome of A. oris MG-1, formerly known as Actinomyces naeslundii MG-1 (Mishra et al., 2010), revealed three sortase genes – one encoding the housekeeping sortase, previously named SrtA (Mishra et al., 2007) that apparently belongs to the class E sortase enzymes (Spirig et al., 2011, Dramsi et al., 2005, Comfort & Clubb, 2004), and two others encoding class C sortases SrtC1 and SrtC2; the latter are involved in the assembly of two distinct forms of fimbria displayed on the Actinomyces cell surface (Mishra et al., 2007). SrtC1 catalyzes formation of type 1 fimbriae that mediate Actinomyces interaction with host cell surface via fimbrial binding to proline-rich proteins coating the tooth surface (Wu et al., 2011). SrtC2, on the other hand, is essential for the assembly of type 2 fimbriae, which are required for bacterial adherence to oral streptococci and host cells via fimbrial interaction with polysaccharide receptors (Wu et al., 2012, Mishra et al., 2007, Mishra et al., 2010). Due to the lack of adequate genetic tools until now, the genetic interrogation of the housekeeping sortase gene srtA in A. oris had been challenging somewhat. In this study, we developed additional genetic tools for this organism and report the unexpected finding that srtA is an essential gene. Deletion of srtA results in bacterial lethality and this lethality is linked to a hitherto unknown glycosylation pathway. This novel finding should make it possible to use A. oris as a convenient in vivo model for identification of sortase inhibitors, which may facilitate the development of a novel class of new antibiotics.
RESULTS
The housekeeping sortase of Actinomyces oris is essential for bacterial viability
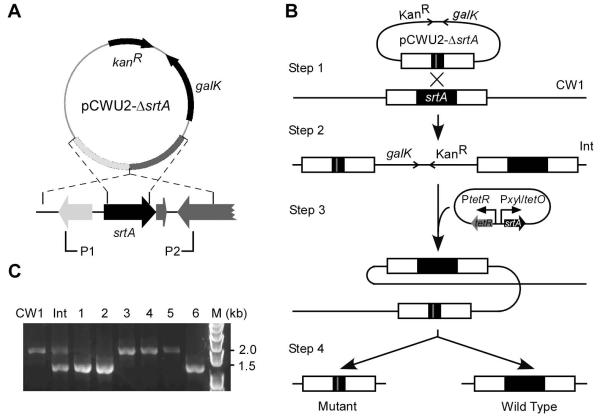
To investigate the role of the housekeeping sortase SrtA in cell surface biogenesis in A. oris, we sought to inactivate the srtA gene by introducing a non-polar, in-frame deletion mutant, using a robust gene deletion method recently established in our laboratory that was successfully used to inactivate the pilus genes as well as two pilus-specific sortases harbored by this bacterium (Mishra et al., 2010). One-kilobase DNA fragments flanking srtA were cloned into the non-replicating vector pCWU2, which expresses galactokinase (GalK) as a counter-selectable marker (Fig. 1A). The resulting plasmid for allele exchange was transferred into a ΔgalK MG-1 derivative, which gave rise to srtA/ΔsrtA merodiploid cells resulting from single-crossover integration of the plasmid. Cells that had undergone a second crossover to expel the plasmid should retain either the wild-type srtA allele or the ΔsrtA mutant allele (Fig. 1B). These were isolated by growing merodiploid cultures without selection for plasmid and selecting rare survivors on agar plates that contained 2-deoxy-D-galactose (2-DG), a metabolite which is converted to toxic 2-deoxygalactose-1-phosphate for cells that retained plasmid-encoded galactokinase. Over 200 survivor colonies were screened and all of them contained the wild-type allele. This surprising result raised the possibility that srtA is an essential gene of A. oris, a feature so far unprecedented for any other Gram-positive organism that has been analyzed to date.
Figure 1.
Genetic disruption of the housekeeping sortase srtA in A. oris. (A) Presented is the non-replicative vector pCWU2 used to generate unmarked, in-frame gene deletions in A. oris. The vector carries 1-kb flanking regions of srtA, the kanamycin resistance cassette (kanR), and galK. Brackets indicate regions amplified by a primer pair P1 and P2. (B) Shown is the schematic of srtA allelic replacement using pCWU2-ΔsrtA. Integration of pCWU2-ΔsrtA into the A. oris ΔgalK mutant CW1 generates a srtA/ΔsrtA merodiploid strain (Int). Excision of srtA from the Int chromosome is permitted in the presence of a plasmid expressing SrtA under the control of a tetracycline-inducible promoter, resulting srtA mutant and wild-type alleles. (C) Excision of srtA from the chromosome of mutant candidates is confirmed by PCR-amplification using the primer pairs P1 and P2.
If our hypothesis that the housekeeping sortase is an essential bacterial product in Actinomyces were correct, it should be possible to delete the WT srtA allele from a srtA/ΔsrtA merodiploid strain that contains an ectopic copy of functional srtA. This turned out to be true: by transforming the srtA/ΔsrtA merodiploid strain with a plasmid in which srtA is constitutively expressed, we were able to delete out the wild type srtA gene from the MG-1 chromosome along with the integrated galK plasmid. To facilitate our functional studies, we next created a conditional mutant of srtA (Fig. 1C), using a newly designed plasmid vector in which srtA expression is tightly regulated under the control of a tetracycline-inducible promoter and a theophylline-responsive riboswitch (pTetR-Ω-SrtA) (see Materials and Methods).
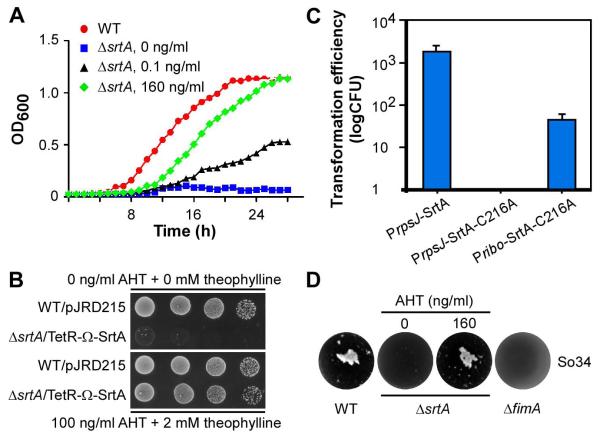
To demonstrate directly that srtA is required for cell viability, we monitored growth rates by measuring optical density of cultures of MG-1 harboring an empty vector and the srtA conditional mutant derivative of MG-1 in a standard broth that was supplemented with various concentrations of inducers anhydrotetracycline (AHT) and theophylline. The srtA mutant was able to grow in the presence of AHT and theophylline, which induced SrtA expression, but not without them (Fig. 2A and see Fig. 4B). In parallel, serially diluted aliquots of the same set of cell cultures were spotted on agar plates with or without AHT and theophylline. The fact that the srtA mutant cells were able to grow when inducers were present but not in their absence proved that the housekeeping sortase is essential for Actinomyces viability (Fig. 2B).
Figure 2.
The housekeeping sortase srtA of A. oris is an essential gene. (A) A conditional mutant of srtA (ΔsrtA) was generated by deleting the chromosomal srtA gene in the presence of the plasmid pTetR-Ω-SrtA expressing SrtA under the control of a tetracycline-inducible promoter in combination with a riboswitch element. Growth of this mutant in the absence (square) or presence of anhydrotetracycline (AHT; 0.1 ng/ml, triangle or 160 ng/ml; diamond) and 2 mM theophylline was monitored by optical density (OD600) and compared to that of the wild-type MG1 strain harboring an empty vector (pJRD215; circle). (B) Ten-fold serial dilutions of overnight cultures of the wild-type MG1 and the conditional ΔsrtA strains in A were spotted on agar plates with and without inducers AHT and theophylline. Cell growth was recorded after 3 days of incubation at 37°C with 5% CO2. (C) Individual plasmids expressing SrtA or SrtA-C216A were electroporated into the MG1 strain. PrpsJ indicates that expression of SrtA is under the control of the strong rpsJ promoter, whereas Pribo refers to a weaker promoter. Transformation efficiency was determined as colony-forming unit (CFU) per microgram of DNA. The results are presented as an average of three independent experiments; each done in triplicates. (D) Interbacterial interaction between S. oralis 34 and the wild-type MG1, fimA deletion mutant, or srtA depleted mutant strain was determined by coaggregation assays.
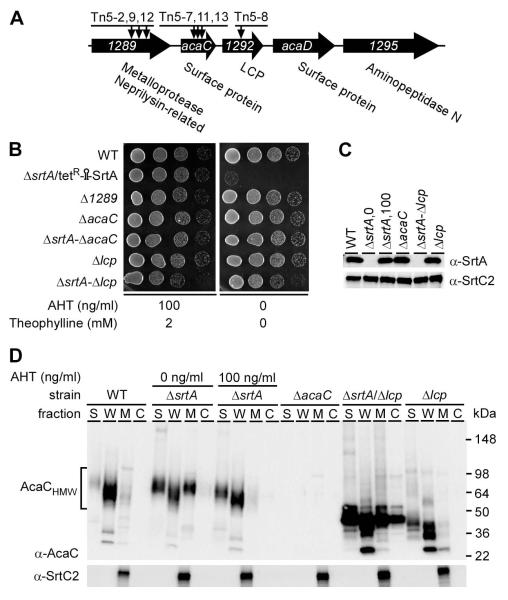
Figure 4.
acaC and lcp are genetic suppressors of srtA. (A) Shown is a graphic representation of the acaC gene locus in the chromosome of A. oris MG1. This locus encodes two surface proteins, a metalloprotease, an aminopeptidase, and a LytR-CpsAPsr (LCP) family protein. Arrows indicate the insertion sites mapped in Tn5 mutants. (B) Cell growth of the MG1, conditional srtA deletion mutant, and various non-polar deletion mutant strains was observed on agar plates in the presence or absence of inducers as described in Fig. 1B. (C) Membrane fractions of various A. oris strains were collected and subjected to western blotting with antibodies against SrtA (α-SrtA) or SrtC2 (α-SrtC2). Numbers 0 and 100 indicate the concentration (ng/ml) of AHT supplemented in the culture of the conditional srtA mutant. (D) Cells of various strains grown to mid-log phase were normalized by optical density. For the conditional srtA deletion mutant, cells were grown in HI broth containing 1% AHT before subcultured in fresh media without (0 ng/ml) or with (100 ng/ml) AHT and 2 mM theophylline. Culture supernatant (S), cell wall (W), membrane (M), and cytoplasmic (C) fractions were obtained by cell fractionation. Equivalent protein samples were separated on 4-12% Tris-Glycine gradient gels and detected by immunoblotting with antibodies against AcaC (α-AcaC) and SrtC2. Molecular mass markers in kDa and high molecular weight AcaC proteins are shown.
Since sortase activity requires a conserved catalytic cysteine residue (Ton-That et al., 2002, Marraffini et al., 2006), we examined whether a SrtA mutant that has the catalytic cysteine residue 216 replaced by alanine is incapable of sustaining bacterial growth. If so, sortase function will be implicated as essential for bacterial viability. Therefore, MG-1 cells were transformed with a plasmid expressing wild-type SrtA or the isogenic C216A mutant, both under the control of a strong promoter (PrpsJ) (Wu & Ton-That, 2010). Transformed cells were plated on agar plates containing kanamycin, and transformation efficiency per microgram of DNA was determined. Remarkably, no transformants were obtained with cells carrying PrpsJ-SrtA-C216A, as compared to PrpsJ-SrtA, which produced normal amounts of transformants (Fig. 2C). We then tested the idea that an excessive expression of the SrtA-C216A mutant might out-compete the endogenous wild-type enzyme in MG-1, thus displaying a dominant negative effect on cell growth. Indeed, compared to no colonies obtained with the PrpsJ-SrtA-C216A plasmid, a measurable number of transformants were observed when MG-1 was transformed with Pribo-SrtA-C216A plasmid (Fig. 2C) (Topp et al., 2010), which expresses a reduced level of SrtA-C216A compared to PrpsJ-SrtA-C216A plasmid.
Housekeeping sortase of A. oris is essential for interbacterial interactions with Streptococcus oralis
A. oris is a key catalyst in the oral biofilm development by virtue of its ability to aggregate with Gram-positive bacteria such as Streptococcus oralis, one of several initial colonizers of the biofilm, and the Gram-negative anaerobe Fusobacteriaum nucleatum, the bridging bacterium that attracts many late colonizers to the biofilm community. It is likely that the housekeeping sortase of A. oris plays a pivotal function in these interactions by catalyzing the cell surface display of specific adhesins that mediate the interbacterial interactions. To address this hypothesis, we next examined whether srtA depletion affects the specific cell-to-cell interaction (i.e. coaggregation) between A. oris and S. oralis, which is known to be mediated by the A. oris fimbrial shaft FimA of type 2 fimbriae (Mishra et al., 2010). To monitor bacterial coaggregation visually, equal numbers (based on optical density) of SrtA-depleted A. oris and standard S. oralis (So34) cells were mixed together in coaggregation buffer and allowed to form co-aggregates, as previously described (Mishra et al., 2010). Compared to MG-1 (WT), SrtA depleted cells failed to co-aggregate with S. oralis, comparable to the deficiency of the fimA deletion mutant (Fig. 2D). Clearly, the housekeeping sortase plays essential roles both in cell growth and in bacterial coaggregation.
Depletion of sortase SrtA affects bacterial septation, envelope morphology, and susceptibility to antibiotics
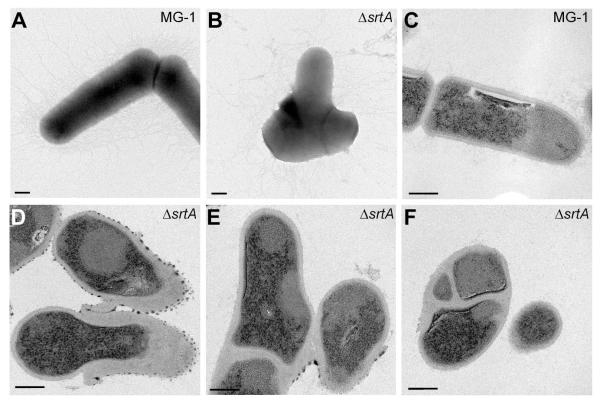
As a step toward understanding why SrtA depletion causes lethality of A. oris, we first analyzed the ultrastructure of SrtA-depleted bacterial cells. Upon culturing bacteria in medium without the inducers of SrtA expression from the complementing plasmid, which resulted in an effective depletion of SrtA as measured by western blots (see below), cells were harvested and directly stained with uranyl acetate, or thin-sectioned prior to negative staining, and viewed by an electron microscope. Remarkably, upon SrtA depletion, the mutant cells displayed some striking morphological aberrations including shorter cell length and bulging (Fig. 3; compare A and B). Electron microscopic analysis of thin-sections revealed that the SrtA-depleted cells also had an unusual cell envelope structure and abnormal division septa (Fig. 3; compare representative structures in C-F).
Figure 3.
SrtA depletion alters cell morphology and displays thickened cell wall and aberrant division septa. Wild-type MG-1 (A and C) and srtA depleted cells (B, D-F) were examined by whole cell (A and B) and thin-section (C-F) electron microscopy. Scale bars indicate 200 nm (A and B) and 100 nm (C-F).
The above data indicate that SrtA-depletion affects the cell envelope. This led us to speculate that an alteration in the cell envelope may lead to a differential sensitivity to antibiotics that target the bacterial cell wall and the membrane, such as vancomycin and daptomycin. Vancomycin binds to the D-Ala-D-Ala moiety of the cell wall precursors preventing cell wall biosynthesis, while daptomycin targets the cell membrane. To quantify antibiotic sensitivity, we employed the Epsilometer test (Etest) (Joyce et al., 1992) and determined the minimal inhibitory concentration (MIC) of vancomycin, daptomycin, and penicillin. Etest strips (bioMerieux) for each antibiotic were placed on a lawn of MG-1 or the srtA conditional mutant expressing SrtA under the control of the TetR promoter in the presence of decreasing concentrations of AHT. Elliptical zones of inhibition were observed and MIC values were recorded (Table 1). Consistent with what was expected, the SrtA-depleted cells were more sensitive to vancomycin and daptomycin as compared to MG-1 cells, with the MICs for each antibiotic being significantly reduced. In contrast, cells with sufficient levels of SrtA expression that sustained normal growth had similar MICs to MG-1. By comparison with vancomycin and daptomycin, A. oris cells appeared to be highly sensitive to penicillin regardless of the status of srtA.
Table 1.
Minimal Inhibitory Concentration (MIC) of penicillin, vancomycin and daptomycin for growth of wild-type and srtA mutant bacteria
| Actinomyces oris strain | MIC (μg ml−1)a |
|||
|---|---|---|---|---|
| Vancomycin | Daptomycin | Penicillin | ||
| MG-1 |
2.03 ± 0.45b | 4.00 ± 1.73b | <0.016 | |
| ΔsrtA/ pTetR-Ω-SrtA |
0 ng/ml AHT | 1.08 ± 0.29b | 1.67 ± 0.29b | <0.016 |
|
|
||||
| 10 ng/ml AHT | 1.37 ± 0.13 | 3.17 ± 0.29 | <0.016 | |
|
|
||||
| 100 ng/ml AHT | 2.45 ± 0.58 | 3.33 ± 0.58 | <0.016 | |
The MICs were determined with Etest stripes as described in the Experimental Procedures. Mean values and standard deviations were calculated from three independent experiments.
P <0.05
Genetic suppressors of srtA deficiency
Conceivably, srtA might be essential because of its requirement for the surface display of one or more cell wall anchored proteins that perform some vital function for Actinomyces. To investigate this logical scenario, we subjected the MG-1 genome to BLAST analysis uncovering a total of 14 putative cell wall anchored proteins each harboring a cell wall sorting signal (CWS), which we previously named AcaA-N (aca for Actinomyces cell wall anchored protein) (Reardon-Robinson et al., 2014). We then deleted each gene using the allele exchange approach earlier described hypothesizing that only nonessential deletion mutants would be generated. To our surprise, all 14 genes were deleted (Reardon-Robinson et al., 2014), demonstrating that none of these presumed SrtA substrates are essential for Actinomyces viability.
Why is SrtA depletion lethal if none of the bacterial products that SrtA sorts to the cell wall are essential? One possibility is that the absence of sortase SrtA might hinder bacterial growth by inducing a toxic phenomenon caused by defective cell wall metabolism. We sought to uncover this underlying process using an unbiased transposon mutagenesis approach. To isolate extragenic suppressors of lethality induced by SrtA depletion, we performed a Tn5 transposon mutagenesis screen for random insertions in the genome that rescues the mutant phenotype. The conditional ΔsrtA mutant harboring the inducible srtA plasmid (pTetR-Ω-SrtA) was mutagenized with a Tn5 transposon carrying a kanamycin (kan) marker. Suppressor mutants were selected for their ability to grow in the absence of AHT and theophylline. Out of roughly 14,000 Tn5 mutants, estimated by counting kan-resistant colonies obtained on plates containing AHT, we obtained a total of 13 kan-resistant survivors, named Tn5-1 to Tn5-13. Except for Tn5-3, which still exhibited abnormal cell morphology, all other Tn5 mutants displayed the normal wild-type cell shape (Fig. S1). Chromosomal DNA of the mutants was then isolated, and genes disrupted by Tn5 were identified by a combination of TAIL-PCR (Liu & Whittier, 1995) and DNA sequencing. The results revealed five classes of suppressors (labeled 1-5), containing transposon insertions within individual genes or a specific gene locus (Table 2).
Table 2.
Genetic suppressors of srtA deletion
| Group | SrtA Suppressors | Targeted Genes | Homologs |
|---|---|---|---|
| Tn5-2, Tn5-9, Tn5-12 | ANA_1289; (2184)a | Metallopeptidase | |
| 1 | Tn5-7, Tn5-11, Tn5-13 | acaC; (963) | Surface protein |
| Tn5-8 | ANA_1292; (1113) | LytR/CpsA family | |
| 2 | Tn5-1 | lepB; (990) | Signal peptidase I |
| 3 | Tn5-3 | ANA_2324; (2439) | Type IV VirB4 component |
| 4 | Tn5-4, Tn5-10 | ANA_2669; (489) | Conserved protein |
| 5 | Tn5-5, Tn5-6 | ANA_0694; (2490) | P-type ATPase |
The number in parenthesis indicates gene length in nucleotides.
Group 1 suppressors harbored transposon insertions within genes encoding a metallopeptidase (ANA_1289), the cell wall anchored protein AcaC (ANA_1291), and a predicted member of the LytR-CpsA-Psr (LCP) protein family (ANA_1292). LCP was initially identified in Bacillus subtilis as a transferase enzyme proposed to catalyze the final step in the cell wall biosynthesis pathway (Kawai et al., 2011). The acaC and lcp genes are part of a locus that also encodes another cell wall anchored protein named AcaD as well as aminopepidase N (see Fig. 4A). Groups 2 and 3 resulted from a single Tn5 insertion into a lepB-like gene and a gene coding for a type IV pilus VirB4-like component, respectively. Finally, the suppressor group 4 represented two independent mutations of the gene encoding a conserved hypothetical protein, whereas the last group was mapped to a gene encoding a P-type ATPase (Table 2). The relation among all of these genes, if any, is not clear to us as yet, and will be probed in future studies. Here, we report the results of several additional lines of experiments that were directed toward AcaC and LCP for the reasons described below.
Excess accumulation of a glycoprotein in the plasma membrane causes the loss of bacterial cell viability in the absence of surface anchoring sortase
Intriguingly, a number of independently isolated Tn5 mutant suppressors affected several adjacent genes (ANA_1289, acaC, and lcp) in the acaC genetic locus (Fig. 4A). This observation led us to focus on characterizing the connection of the affected gene products in lethality induced by SrtA depletion. We reasoned that if suppression of lethality was the result of specific gene disruption by Tn5, then a directed deletion of the respective genes should also have the same suppressor phenotype. Therefore, we generated non-polar, in-frame deletion mutants of ANA_1289, acaC, and lcp, and asked if a double mutant of ΔsrtA could be constructed with any of these mutants (Fig. 4A). Indeed, while we could not obtain a double deletion mutant of srtA and ANA_1289, we succeeded in generating a standard srtA deletion mutant in the absence of either acaC or lcp (Fig. 4B & 4C). Unlike the srtA conditional mutant, which failed to grow in the absence of inducers (Fig. 2A & 2B), the two generated double deletion mutants did not exhibit any growth defects as compared to the parental strain MG-1 (Fig. 4B), confirming that a loss of either acaC or lcp can act as a SrtA bypass suppressor. Importantly, deletion of acaC or lcp gene by itself did not affect cell growth (Fig. 4B) or SrtA expression (Fig. 4C). By RT-PCR and Western blot analyses, we found that all three Tn5 mutants Tn5-2, Tn5-9, and Tn5-12, each of which suppressed lethality induced by SrtA-depletion, had drastically reduced acaC expression (see Figures S2 & S3). Thus, it is likely that Tn5 insertion of ANA_1289 in the three Tn5 mutants did not suppress the lethality associated with srtA deletion, but rather caused a polar effect to the adjacent gene acaC.
Toxic accumulation of AcaC in membrane
To directly monitor the expression and the subcellular location of native AcaC protein by immunoblotting, we generated polyclonal antibodies against recombinant AcaC protein lacking the N-terminal signal peptide and C-terminal CWS domain. Cells grown to mid-log phase were subjected to fractionation and protein samples in each cellular fraction were separated by SDS-PAGE, followed by immunoblotting with specific antibodies. As expected, AcaC was detected in the cell wall fraction of MG-1 (Fig. 4D; lanes WT). Surprisingly, although the AcaC precursor protein is predicted to have a molecular mass of 32 kDa, AcaC protein detected by its antibody migrated as a smear between 50- and 98-kDa markers, suggestive of protein glycosylation. Intriguingly, when srtA expression was repressed, AcaC protein accumulated in the membrane fraction (Fig. 4D; lanes ΔsrtA, 0 ng/ml). In sharp contrast, this membrane accumulation of AcaC was alleviated to wild-type levels when SrtA expression was induced (Fig. 4D; lanes ΔsrtA, 100 ng/ml). As a control for specificity of the anti-AcaC antibody, no protein band was detected in the mutant that lacks acaC (Fig. 4D; lanes ΔacaC). Intriguingly, in the srtA and lcp double mutant, most of the high molecular weight AcaC bands (~50-100 Kda AcaChmw) were not detected, while several low molecular weight AcaC bands (~30-55 kDa) were observed (Fig. 4D; lanes ΔsrtA/Δlcp). The lower molecular weight AcaC bands were also produced in the absence of lcp alone, and these antigens were mostly anchored to the cell wall with some secreted into the culture medium but very little in the membrane (Fig. 4D; lanes Δlcp). This result suggests that LCP may be involved in some kind of post-translational modification of AcaC.
AcaC is glycosylated in a LCP-dependent pathway
One possibility for the production of high molecular weight AcaC protein species with a smeary gel mobility pattern could be that AcaC is glycosylated. To investigate this, we engineered an AcaC protein that contains a Tobacco Etch Virus (TEV) cleavage site, followed by 6-histidine tag inserted just upstream of the AcaC cell wall sorting signal (Fig. 5A). The recombinant protein was expressed in the A. oris mutant ΔacaC and purified by affinity chromatography from the cell wall fraction after digestion of peptidoglycan. Glycosylation of purified AcaC proteins was detected by the periodic acid-Schiff (PAS) method using a glycoprotein staining kit (Pierce). As shown in Fig. 5B (left panel-lane 1), two major AcaC species were purified from A. oris; one band migrating around 49 kDa and another somewhat smeary band migrating from 64 to 115 kDa. The higher molecular weight band reacted strongly to PAS, while the ~49 Kda band was barely cross-reactive (Fig 5B- compare lane 1 in left and right panels). The control recombinant protein AcaC purified from E. coli, which lacks an N-terminal signal peptide and C-terminal CWS, was negative for PAS staining (Fig. 5B, third lanes). Western blotting with antibodies against AcaC (i.e. α-AcaC) showed that both species are AcaC derived (Fig. 5C; second lane). Noticeably, the 49 kDa species exhibited similar mobility to that of the AcaC species isolated from the culture medium and cell wall fractions of the double mutant ΔsrtA-Δlcp and single mutant Δlcp, respectively (Fig. 5C; compare the second lane to the fourth and fifth lanes). In addition, the high molecular weight AcaC species were not detected in these mutants. We conclude that AcaC is subject to two distinct modification events, one of which is dependent on LCP. The molecular basis of the other modification step that produces the ~49 Kda band remains to be elucidated.
Figure 5.
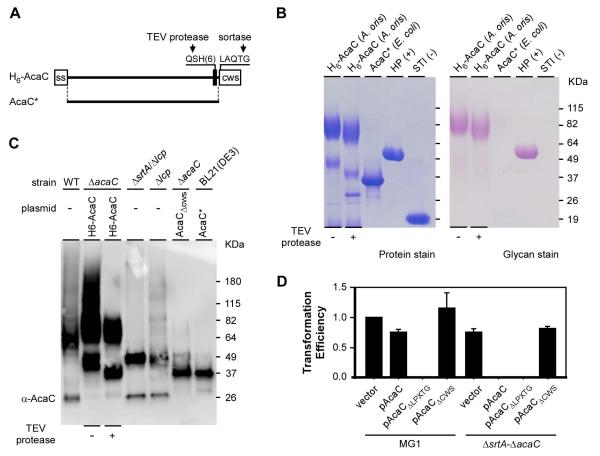
AcaC is a cell wall anchored glycosylated protein. (A) Shown is a diagram of a recombinant AcaC protein with its signal sequence (ss), TEV cleavage site followed by a six-histidine tag, and cell wall sorting signal (CWS) with the LAQTG motif. A truncated version of AcaC lacking the signal sequence (ss) and CWS is denoted as AcaC*. (B) Purified AcaC proteins from E. coli and A. oris were separated by SDS-PAGE and stained with Coomassie (protein stain) or Periodic acid–Schiff (glycan stain). Nonglycosylated soybean trypsin inhibitor (STI) and glycosylated horseradish peroxidase (HP) were used as controls. (C) The His-tagged AcaC proteins purified from A. oris in B were treated with TEV protease and subjected to immunoblotting with α-AcaC; their mobility was compared with AcaC species isolated from cell wall fractions of the MG1 (WT) and Δlcp mutant strains, culture medium fractions of mutants ΔsrtA/Δlcp and ΔacaC/pAcaCΔcws, and truncated AcaC purified from E. coli BL21(DE3). (D) Transformability of the empty vector and plasmids expressing full-length AcaC (pAcaC), AcaC lacking the LPXTG motif (ΔLPXTG), or AcaC lacking the CWS in strains MG1 and its isogenic mutant ΔsrtA-ΔacaC was determined by cell growth as described in Fig. 3B. The transformation efficiency values, averages of three independent experiments, are presented relative to that of the empty vector in MG1, which was arbitrarily assigned as 1.
To localize glycosylation sites within AcaC, purified H6-AcaC proteins were treated with TEV to remove the protein C-terminus (see Fig. 5A) prior to separation on SDS-PAGE for Western blotting and glycan staining. After TEV treatment, the 49 kDa species was trimmed down to roughly 37 kDa and barely visible by PAS staining, while the large molecular weight species were only slightly reduced in mass but still strongly reactive to the glycan stain (Fig. 5B). By Western blot analysis, AcaC was detected in both species (Fig. 5C, third lane). Together, these results show that the LCP-dependent glycosylation is localized to the N-terminal segment of AcaC preceding the CWS.
Membrane accumulation of glycosylated AcaC is toxic
Lethality of srtA deletion might be the consequence of an excessive accumulation of the glycosylated AcaC protein in the membrane, which is normally destined for the cell surface but gets trapped in the membrane in the absence of SrtA (see Discussion, Fig 6). If this is the case, we wondered whether a secreted form of AcaC (AcaCΔCWS) would no longer be lethal in the absence of srtA. To test this hypothesis, we generated recombinant plasmids expressing a full-length AcaC (pAcaC), a mutant AcaC devoid of the LPXTG motif (pAcaCΔLPXTG), or a mutant AcaC lacking the entire CWS (pAcaCΔCWS). Individual plasmids were transformed into MG1 or the ΔsrtA-ΔacaC mutant, and transformation frequency was calculated (Fig. 5D). While the transformation frequency of pAcaC with MG1 cells was roughly 20% less than that with the empty vector, a value that was within experimental variation, no transformation was observed with pAcaC in the ΔsrtA-ΔacaC mutant cells, as expected. Likewise, no transformants were obtained with pAcaCΔLPXTG, a mutant that is retained within the membrane. In sharp contrast, the transformation frequency of pAcaCΔCWS, which expresses AcaC lacking the membrane localization domain, was comparable to that of the empty vector (Fig. 5D; last lane). We conclude that in the absence of sortase to catalyze cell wall anchoring of surface proteins, the excessive membrane accumulation of a glycosylated cell surface protein perturbs the cell envelope sufficiently to block bacterial cell viability.
Figure 6.
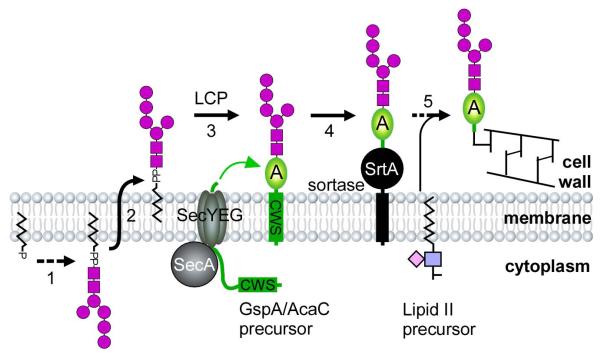
A model of a glycosylation pathway for AcaC, renamed as GspA (gsp for glycosylated surface-linked protein), in A. oris is proposed. Glycan precursors are synthesized in the cytoplasm (Step 1), transported across the cytoplasmic membrane (Step 2), and serve as substrates for LCP. Functioning as a glycosyltransferase, LCP mediates attachment of the glycans to a membrane-bound GspA precursor (Step 3). The housekeeping sortase SrtA then catalyzes cell wall anchoring of glycosylated GspA (Step 4) via the lipid II precursor (Step 5).
DISCUSSION
A unique feature of cell envelope biogenesis in Gram-positive bacteria is the covalent attachment of many critical cell surface proteins to the peptidoglycan layer that encases the plasma membrane. As described in the introduction, a transpeptidase enzyme known as sortase catalyzes this covalent joining of peptidoglycan with specific designated proteins that harbor a cell wall sorting signal. Most Gram-positive bacteria harbor a class A type sortase (SrtA) that is referred to as the housekeeping sortase because a large battery of surface proteins encoded by an organism are the dedicated substrates of this single transpeptidase (Comfort & Clubb, 2004, Dramsi et al., 2005). The housekeeping sortase serves as a major virulence factor since many of its substrate cell surface proteins play important roles in bacterial pathogenesis (Marraffini et al., 2006). Significantly, genetic disruption of SrtA affects pathogenesis of several Gram-positive pathogens that have been characterized prior to this study, but not viability of any of these pathogens. In this context, we report here the surprising discovery that SrtA is an essential function in Actinomyces oris, which is one of the major players in the initiation of a multi-bacterial biofilm called dental plaque.
To demonstrate the predicted role of A. oris SrtA in the multi-bacterial biofilm development, we sought to disrupt the SrtA gene using a facile allele exchange methodology for this organism that was recently developed in our lab. Surprisingly, several attempts of allele exchange produced recombinants that always retained the wild type allele of srtA (Fig. 1), suggesting that this gene might be essential for the viability of A. oris. This inference was correct: we were able to create a conditional srtA knock-down mutant, which is viable under induced conditions (AHT+Thio- containing medium) but loses viability in the absence of inducers. We then went on to show that SrtA depletion is detrimental to cell growth and morphology (Figures 1 and 2). This is likely due to some perturbations in the cell envelope that cause an increased sensitivity of the mutant cells to vancomycin and daptomycin under repressive conditions that lead to SrtA depletion (Table 1). Finally, as expected, A. oris cells with SrtA depletion failed to display cell surface adhesins and was also defective in co-aggregating with S. oralis (Fig. 2). This establishes the important role of A. oris SrtA in a specific cell-cell interaction that underlies the oral biofilm development.
A key question that arose is how the depletion of the housekeeping sortase affects the cell envelope and in turn, the viability of Actinomyces. We speculated that the loss of viability in this unusual case is due to the absence of a surface protein that plays some kind of an essential role in the growth and viability of A. oris. Therefore, we deleted genes encoding each of the 14 predicted surface proteins expressed by this organism. To our surprise, we succeeded in generating all the individual deletion mutants, suggesting that the lethality is caused by some other mechanism. Based on our evidence that the sortase-depleted cells have some type of alteration in the cell envelope, a logical hypothesis was that the absence of sortase might be toxic. In this light, we thought that the established approach of random transposon mutagenesis might help to disrupt the responsible toxic gene product and as a result, rescue the viability of A. oris ΔsrtA. To that end, we performed Tn5 transposition on the conditional srtA deletion chromosome and screened for survivors that formed colonies without inducers. We succeeded in isolating five groups of suppressors that eliminated ΔsrtA lethality (Table 2).
A striking feature of the battery of srtA bypass suppressors mutants we obtained is that a number of these mapped to one of several genes at a chromosomal locus that encodes a cell wall anchored protein (we previously designated as AcaC) and a LCP-like protein (Figures 4 and 5). Using the acaC and lcp single deletion mutants, we obtained double deletion mutants of acaC and srtA or lcp and srtA, respectively, thereby demonstrating that acaC and lcp are genetic bypass suppressors of srtA. Curiously, further work revealed a smeary appearance of AcaC antigen in SDS-PAGE analysis, leading us to suspect and subsequently prove that AcaC is a glycoprotein. Prompted by the genetic proximity of AcaC and lcp, we wondered whether LCP (and possibly some other unlinked genetic suppressors) might be involved in the various steps of the AcaC glycosylation pathway.
In support of the conjecture that LCP-like proteins may possess glycosyl transferase activities (Kawai et al., 2011, Chan et al., 2013), deletion of lcp was associated with the disappearance of the smeary, high molecular weight antigenic forms of AcaC and the concomitant appearance of a uniform band consistent with the predicated molecular mass of the non-glycosylated AcaC protein (Figures 4 and 5). We therefore propose that A. oris LCP catalyzes the final step of glycosylation by linking glycan precursors to AcaC prior to cell wall anchoring. While this hypothesis and the glycosylation pathway remains to be molecularly characterized and dissected, it is important to note that AcaC contains several NXT/S motifs that may be subject to glycosylation at the serine or threonine residue (Schwarz & Aebi, 2011).
How does the absence of AcaC or the presumed glycosyl transferase (LCP) that glycosylates it rescue effects of srtA depletion? We propose that excess accumulation of the AcaC glycoproteins impart envelope stress that causes growth arrest and ultimately cell death. Furthermore, the toxicity must be ascribed to the glycosyl moiety (“glyco-stress”) since accumulation of unmodified AcaC in the absence of SrtA and LCP is not lethal. Because AcaC glycosylation is most likely a part of an assembly line-like process of cell envelope assembly, which must couple protein secretion and SrtA-mediated cell wall anchoring, it is tempting to speculate that the block in the transfer of the glycoprotein from the membrane to the cell wall in the absence of SrtA results in a “membrane-jamming” phenomenon due to excess accumulation of glycosylated AcaC precursors in the cytoplasmic membrane. The fact that AcaC molecules that lack the CWS were non-glycosylated and secreted into the culture medium, and no longer caused a lethal phenotype gives credence to this hypothesis (Fig. 5). Further consistent with our hypothesis is the isolation of three acaC suppressor mutants [Tn5-7, Tn5-11, and Tn5-13 (Table 2)], in which Tn5 insertion landed at nucleotide positions 340, 477, and 479 of the 963 base-pair acaC gene, respectively, each of which truncated the C-terminal CWS domain of AcaC. Lastly, it is noteworthy that glycosylation is absent in the AcaCΔCWS mutant (Fig. 5). This supports our hypothesis that glycosylation occurs after AcaC is translocated across the membrane and inserted into the membrane via its CWS.
Intriguingly, loss of LepB could also suppress the lethality of SrtA deficiency in strains that have intact acaC and lcp genes (see Table 2; group 2). How could we explain this result? LepB is a type 1 signal peptidase which removes the signal peptide of protein precursors, permitting the maturation and proper folding of proteins translocated across the bacterial cytoplasmic membrane (van Roosmalen et al., 2004). Since AcaC contains an N-terminal signal peptide, the absence of LepB may leave AcaC unprocessed, which may prevent further membrane insertion. Under this condition, the absence of srtA by genetic disruption was not lethal because AcaC may not be glycosylated. Indeed, no AcaC was detected in the Tn5-1 suppressor (i.e. lepB and srtA mutant; Table 2 and Fig. S3). It is logical to question why LepB deficiency itself is not lethal in Actinomyces when lepB is an essential gene in E. coli (Dalbey & Wickner, 1985). We posit that the deletion of lepB in A. oris is most likely not lethal because this organism harbors another type 1 signal peptidase, whose gene [ANA_1188] is located upstream of lepB [ANA_1190]. It is likely that LepB is singularly most important for AcaC processing, but this remains to be confirmed.
While the involvement of AcaC, LCP and LepB in the lethal phenotype of srtA deletion is apparent, the mechanisms of suppression for the remaining groups of suppressors are not understood (Table 2). Among these is ANA_2324, which encodes a homolog of VirB4, an ATPase which might be potentially involved in substrate translocation across the membrane (Bhatty et al., 2013). ANA_2669 is the last gene of a locus that encodes an ABC-type transport system. While the ANA_2669 protein is conserved, it has not been ascribed with a function yet. Finally, the protein encoded by ANA_0694 is a P-type ATPase, which belongs to a large superfamily of cation and lipid pumps, some members of which are phospholipid flippases (Palmgren & Nissen, 2011). Interestingly, all insertion mutations of ANA_2324, ANA_2669, and ANA_0694 in the absence of srtA (i.e. Tn5-3, Tn5-10, and Tn5-5), resulted in the membrane localization and accumulation of heterogenous glycosylated forms of AcaC, whereas in the other suppressor of group 4 (i.e. Tn5-4), no glycosylated AcaC antigen were observed (Fig. S3). Altogether, the results suggest that these proteins may be involved in the metabolism and translocation of glycan precursors across the membrane. Further genetic and biochemical work will be needed to illuminate the AcaC glycosylation pathway. This is an important future direction in investigating Gram-positive envelope assembly. Since glycosylated AcaC likely acts as a glycoprotein adhesion whose pathogenic or commensal function has yet to be unveiled, we prefer to rename this protein as GspA (for glycosylated surface linked protein A). Significantly, GspA homologs are found in most Actinomyces species sequenced to date, suggesting that the GspA glycosylation pathway and SrtA essentiality are conserved in the Actinomyces genus.
We conclude by proposing the following model of GspA glycosylation coupled to sortase-mediated cell wall anchoring (Fig. 6). GspA precursors are synthesized in the cytoplasm, transported across the membrane by the Sec apparatus, and retained within the secretory pathway by the CWS. Separately, glycan precursors are produced by a different enzymatic pathway, which involves many proteins, including membrane transport systems and LCP. Acting as a glycosyltransferase, LCP joins specific glycan strands to a GspA molecule emerging from the Sec machinery. In the last step, the housekeeping sortase catalyzes the joining of glycosylated GspA molecule to the bacterial peptidoglycan via the lipid II precursor. When SrtA expression is diminished or SrtA is inhibited, GspA continues to accumulate in the membrane and become glycosylated until envelope stress resulting from this prevents cell growth ultimately causing cell death. Furthermore, we envision that inhibitors of sortase should emerge as novel antibiotics. It is noteworthy to point out that efforts have been made to identify inhibitors of sortase, both from natural products and synthetic compounds, using an in vitro FRET assay with recombinant sortase enzymes (Ton-That et al., 2000). Many potent sortase inhibitors have been reported (Maresso & Schneewind, 2008, Maresso et al., 2007), but a general concern is whether these compounds are active in vivo. In this context, it is important to point out the obvious that the Actinomyces system provides a convenient in vivo assay for new antibiotics discovery based on sortase. Lastly, since glycosylation might modulate host immunity by oral bacteria (Settem et al., 2014), the genetic tools we describe for A. oris will permit future studies of this important aspect of oral microbial pathogenesis.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids and media
Bacterial strains and plasmids used in this study are listed in Supporting Table S1 of Supporting Information (SI). Generation of recombinant plasmids was described in Supporting Materials and Methods (see SI). Actinomyces were grown in heart infusion broth (HIB; Fisher Scientific) or on HIB agar plates. Streptococci were grown in Brain Heart Infusion (BHI; Fisher Scientific) supplemented with 0.5% glucose. Escherichia coli strains were grown in Luria broth (LB). When required, antibiotics (kanamycin or streptomycin) were added into medium at a final concentration of 50 μg ml−1. Reagents were purchased from Sigma unless indicated otherwise. Except for antibodies against AcaC (see SI), the generation of other antibodies was described elsewhere (Mishra et al., 2007).
Construction of an inducible gene expression system in A. oris
To prepare the backbone vector pJRD-Sm with a streptomycin resistance gene, the kanamycin resistance gene of pJRD215 (Yeung & Kozelsky, 1994) was excised by reverse PCR with appropriate primer sets (Supporting Table S2) using pJRD215 as template. A DNA fragment encompassing the inducible Pxyl/tetO promoter and tetR amplified from plasmid pRMC2 (Corrigan & Foster, 2009) was fused to the MG-1 srtA gene with its ribosomal binding site (rbs). The resulting fragment was cloned into pJRDSm to produce pTetR-SrtA. Due to leakiness of this tetracycline-inducible system in A. oris, the srtA rbs was replaced by a riboswitch element (E*) (Topp et al., 2010) using reverse PCR with the primer pair Ribo-srtA-F and Ribo-tet-R and pTetR-SrtA as template. The resulting vector pTetR-R*-SrtA was used to construct the tightly inducible vector pTetR-Ω-SrtA, in which the strong, consecutive promoter of fimQ was inserted upstream of the Pxyl/tetO promoter to enhance TetR repressor expression.
Gene deletions in A. oris
Non-polar, in-frame deletion mutants in A. oris were generated using a previously published protocol (Mishra et al., 2010). Briefly, a deletion vector carrying galK as a counterselectable marker (Supporting Table S1) was electroporated into CW1, an isogenic mutant of MG-1 that lacks galK (Mishra et al., 2010). Integration of the vector into the bacterial chromosome via homologous recombination was selected on HIB agar plates containing 50 μg ml−1 kanamycin. Excision of the vector via a second recombination event – which results in gene deletion or reconstitution of the wild-type genetype – was selected by 2-deoxy-D-galactose (2-DG). 2-DG resistant and kanamycin-sensitive colonies were screened for the expected deletion mutation by PCR amplification. Candidate deletion mutants were also confirmed by Western blotting with specific antibodies when available. The same procedure was employed for construction of double deletion mutants.
To generate a conditional deletion mutant of srtA, the deletion plasmid pCWU2-ΔsrtA was electroporated into CW1 and plasmid integration was selected by kanamycin resistance. Integrant cells were then transformed with pTetR-Ω-SrtA, and the resulting transformants were selected on HIB agar plates supplemented with kanamycin and streptomycin. A colony of transformants was used to inoculate an overnight culture in HIB containing two inducers (100 ng ml−1 AHT and 2 mM theophylline). 100-μl aliquots of 100-fold diluted overnight cultures in fresh HIB were spread out on HIB agar plates containing streptomycin and both inducers. After three days of incubation at 37°C and with 5% CO2, the resulting streptomycin-resistant colonies were screened for kanamycin sensitivity. Kanamycin-sensitive colonies were then screened for the loss of the srtA chromosomal copy by PCR amplication. The generated conditional srtA deletion mutant was analyzed for cell growth and SrtA expression by immunoblotting with antibodies against SrtA (α-SrtA).
Tn5 transposition
The vector pMOD-2 <MCS> (Epicentre) was used to prepare the EZ-Tn5 transposon as the following. The kanamycin resistant gene cassette was PCR-amplified using the primer pair Kan-F/R and pJRD215 as template. The PCR product was digested by BamHI and cloned into pMOD-2 <MCS> pre-cut with BamHI to yield pMOD-2/Kan215. The primers ME-9-R (CTGTCTCTTATACACATCTCAACCATCA) and ME-9-F (CTGTCTCTTATACACATCTCAACCCTGA) were used to PCR-amplify the Tn5 transposon from pMOD-2/Kan215.
Production of Tn5 transposome and transposition assays were performed according to protocols provided the manufacturer (Epicentre). Briefly, for transposome production 200 ng of Tn5 transposon DNA was mixed with 4 units of EZ-Tn5 transposase (Epicentre) and incubated for 2 h at room temperature. 1.5 μl of the transposome was mixed with 200 μl the competent cells of the srtA conditional mutant and incubated on ice for 10 min before electroporation. Electroporated cells were spread on HIB agar plates containing both streptomycin and kanamycin. Streptomycin- and kanamycin-resistant colonies were further analyzed by immunoblotting for the absence of SrtA expression.
To characterize the insertion sites by Tn5 transposition, thermal asymmetric interlaced PCR (TAIL-PCR) was performed according to the published protocol(Liu et al., 1995) with some modifications to suit the high GC content of Actinomyces DNA. The first round of PCR amplification was performed using Phusion DNA polymerase (New England Biolabs) with primers Tn5-1 (CGAACTGTTCGCCAGGCTCAAG) and AD1 (NGTCGASWGANAWGAA) – a degenerate primer – and chromosomal DNA isolated from Tn5 mutants. The PCR conditions were similar to the published protocol, except that denaturing temperature was set to 98°C. 1 μl of the first-round PCR products was used as template in the second-round PCR with primers Tn-2 (CTGACCGCTTCCTCGTGCTTTA) and AD1. The PCR products were gel-purified and cloned into a TOPO blunt-end cloning vector (Life Technologies). The cloned PCR products were subjected to DNA sequencing using universal primers M13F/M13R (CCGAGCAGTCTCTGTCCTTC/CCCTCTCACTCCCTTCCTG). Sequences that harbor the unique mosaic end sequence of the Tn5 transposon (AGATGTGTATAAGAGACAG) were considered bona fide Tn5-targeted sites.
Identification of srtA genetic suppressors
The commercially available EZ-Tn5 transposon was modified to carry a kanamycin selectable marker for A. oris using pMOD-2<MCS> (Epicentre). For transposome production, 200 ng of Tn5 transposon DNA was mixed with 4 units of EZ-Tn5 transposase (Epicentre) and incubated for 2 h at room temperature. 1.5 μl of the transposome was mixed with 200 μl the competent cells of the srtA conditional mutant and incubated on ice for 10 min before electroporation. Electroporated cells were spread on HIB agar plates containing both streptomycin and kanamycin. Streptomycin- and kanamycin-resistant colonies were further analyzed by immunoblotting for the absence of SrtA expression.
To characterize the insertion sites by Tn5 transposition, thermal asymmetric interlaced PCR (TAIL-PCR) was performed according to the published protocol (Liu & Whittier, 1995) with some modifications to suit the high GC content of Actinomyces DNA (see SI). PCR products were gel-purified and cloned into a TOPO blunt-end cloning vector (Life Technologies) for DNA sequencing using universal primers M13F/M13R. Sequences that harbor the unique 19-bp mosaic end of the Tn5 transposon were considered bona fide Tn5-targeted sites. Subsequently, candidate suppressors were examined by electron microscopy for cell morphology and Western blotting analysis for protein expression and localization.
Cell growth assays
Cells of the conditional srtA deletion mutant and parental strain MG-1 harboring the empty plasmid pJRD215 were grown overnight in HIB containing AHT (0.1 ng/ml) and streptomycin (50 μg/ml) at 37°C with 5% CO2 in air atmosphere. Harvested cells were washed once to remove AHT and used to inoculate three cultures in fresh HIB (1/200 dilution), each containing streptomycin and various concentrations of AHT (0, 0.1, and 160 ng/ml) and 2 mM theophylline. Cell growth at 37°C was monitored by measuring optical density at 600 nm (OD600) every hour for 30 h. The experiment was repeated at least three times.
For cell growth on plates, washed cells from overnight cultures were normalized to the same OD600, subjected to 10-fold serial dilutions, and spotted on HI agar plates containing with or without AHT (100 ng/ml) and theophylline (2 mM). After incubation at 37°C for 3 days, cell growth was recorded.
Electron Microscopy
To observe cell morphology, A. oris cells grown in HIB agar plates were suspended in 0.1 M NaCl and washed with water. A drop of bacterial suspension in PBS was placed onto carbon-coated nickel grids and stained with 1% uranyl acetate. Samples were examined using a JEOL JEM1400 electron microscope.
For thin-section electron microscopy, harvested cells were fixed overnight at 4°C with 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4. Fixed cells were treated with 0.1% borohydride in Millonig’s buffer, dehydrated in ethanol, and embedded in LR White resin (EMS; Hatfiedl, PA). Ultrathin sections (roughly 50 nm) on grids were stained with 1% uranyl acetate and examined microscopically.
Cell fractionation and western blotting
Cell fractionation and Western blotting were followed according to a published protocol (Wu et al., 2012). Briefly, mid-log phase cultures of A. oris strains were normalized to equal OD600 and cells were fractionated into culture medium (S), cell wall (W), membrane (M) and cytoplasm (C) fractions. Isolated fractions were subjected to protein precipitation by 7.5% trichloroacetic acid, followed by washing with cold acetone. Protein samples were dissolved in hot sodium dodecyl sulfate (SDS)-containing sample buffer, separated by 4-12% Tris-glycine gradient gels, and subjected to immunoblotting with specific antisera (α-SrtA, α-SrtC2 and α-AcaC; 1:100 dilution), followed by chemiluminescence detection.
Periodic Acid-Schiff (PAS) staining
PAS staining was carried out using a glycoprotein staining kit (Thermo Scientific) according to the manufacturer’s protocol. Specifically, purified AcaC proteins from A. oris were first treated with TEV protease or mock-treated. For a 25 μl TEV cleavage reaction, 20 μg AcaC protein was digested with 20 U of AcTEV protease (Invitrogen) in Reaction Buffer containing 4 mM DTT for 12 h at 30°C. After protease treatment, protein samples were subjected to SDS-PAGE electrophoresis in duplicates, with the recombinant AcaC protein purified from E. coli, glycosylated horseradish peroxidase (HP), and nonglycosylated soybean trypsin inhibitor (STI; Thermo Scientific) included as controls. One gel was stained by Coomassie brilliant blue and the other was fixed with 50% methanol. The fixed gel was then treated with 25 ml of oxidizing solution containing periodic acid, which oxidizes the vicinal diols present in glycoproteins to aldehydes. After three 5-min rinses with 3% acetic acid, the gels were soaked in 25 ml of the Glycoprotein Staining Reagent and then in 25 ml of the Reducing Solution for 5 min with gentle agitation. Glycosylated proteins were stained, yielding magenta bands.
Real-time PCR
A. oris cells grown in HI broth to an OD600 of 0.3 at 37°C in a water bath shaker were collected by centrifugation. Cell pellets were suspended in 1 ml of ice-cold Trizol (Sigma) and disrupted with 0.1 mm Zirconia Beads using a mini-bead beater (Biospec products). After centrifugation, supernatants were collected and RNA purified using the Ambion RiboPure™- Bacterial kit according to the manufacturer’s protocol. The extracted RNA samples were treated with DNase I (Ambion) to remove traces of chromosomal DNA. And RNA samples were further cleaned with the Qiagen RNeasy MinElute™ clean-up kit. Total RNA (300 ng) was used for cDNA synthesis using Stratascipt RT (Stratagene) based on the manufacturer’s instruction. Real-time PCR was performed using MyiQ™ single color Real-time PCR Detection System and reactions were prepared using SYBR® Green PCR Master Mix with appropriate primers (Table S1). Changes in gene expression were calculated using the ΔΔCT method as the following, ΔCT = CT (target) -CT (housekeeping gene); ΔΔCT = ΔCT1 - ΔCT2; Fold changes were calculated as 2−ΔΔCt. The 16S rRNA gene was used as the housekeeping gene reference and all samples included reactions without reverse transcriptase as control to assess genomic DNA contamination in the reactions.
Epsilometer test for antibiotic susceptibility
Overnight cultures of the srtA conditional mutant grown in HIB supplemented with AHT (1.0 ng/ml) were harvested by centrifugation, and cells were washed in fresh medium and diluted 100 fold. 100-μl aliquots of cell suspensions were spread on HIB agar plates containing various concentrations of AHT (0, 10, and 100 ng/ml). Each E-test strip containing benzylpenicillin, vancomycin or daptomycin (bioMerieux) was laid on bacterial plates. After 3 days of incubation at 37°C, elliptical zones of inhibition were observed on plates and the minimum inhibitory concentration (MIC) of each antibiotic was recorded. Mean values and standard deviations of MICs were calculated from three independent experiments. Statistical significance was determined by Student’s t test.
Bacterial coaggregation assay
Co-aggregation between A. oris and S. oralis was performed as previously described (Mishra et al., 2011). Briefly, stationary-phase cultures of bacterial strains grown in CAMG complex medium with 0.5% glucose were harvested by centrifugation. Bacterial cells were washed in Tris-buffered saline (TBS, pH 7.5) containing 0.1 mM CaCl2 and normalized to an OD600 of 2.0 (approximately 2 × 109 CFU/ml). 0.4-ml aliquots of Actinomyces and streptococcal cell suspensions were mixed in 24-well plates for a few minutes on a rotator shaker and coaggregation were recorded by an Alpha Imager (Alpha Innotech).
Purification of AcaC from A. oris
The A. oris ΔacaC strain harboring pAcaCH6-TEV was used to inoculate an overnight culture in HIB supplemented with kanamycin, which was then diluted 100-fold in fresh medium and grown until OD600 of 0.7. Cells were harvested by centrifugation, washed twice in sterile water, suspended in 35 ml of SMM buffer (0.5 M sucrose, 10 mM MgCl2 and 10 mM maleate, pH 6.8), and treated with 500 U/ml mutanolysin plus 150 mg lyzosome at 37°C overnight. After treatment, soluble cell wall fractions were separated from the protoplasts by centrifugation and dialyzed against EQ buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5) at 4°C overnight prior to AcaC purification by affinity chromatography. Purified proteins were concentrated by filtration using Amicon-10K (Millipore) and stored at -20°C for Periodic Acid-Schiff (PAS) staining and Western blot analysis.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Timothy James Foster (The University of Dublin, Ireland) for pRMC2. Research reported in this publication was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under Award Numbers F31DE024004 (to M.E.R-R.) and DE017382 (to H.T-T.). The authors have no conflict of interest to declare.
REFERENCES
- Bhatty M, Laverde Gomez JA, Christie PJ. The expanding bacterial type IV secretion lexicon. Res Microbiol. 2013;164:620–639. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Mazmanian SK, Trost M, Pucciarelli MG, Liu G, Dehoux P, Jansch L, Garcia-del Portillo F, Schneewind O, Cossart P. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol Microbiol. 2002;43:869–881. doi: 10.1046/j.1365-2958.2002.02798.x. [DOI] [PubMed] [Google Scholar]
- Bolken TC, Franke CA, Jones KF, Zeller GO, Jones CH, Dutton EK, Hruby DE. Inactivation of the srtA gene in Streptococcus gordonii inhibits cell wall anchoring of surface proteins and decreases in vitro and in vivo adhesion. Infect Immun. 2001;69:75–80. doi: 10.1128/IAI.69.1.75-80.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzik JM, Marraffini LA, Schneewind O. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol Microbiol. 2007;66:495–510. doi: 10.1111/j.1365-2958.2007.05939.x. [DOI] [PubMed] [Google Scholar]
- Chan YG, Frankel MB, Dengler V, Schneewind O, Missiakas D. Staphylococcus aureus mutants lacking the LytR-CpsA-Psr family of enzymes release cell wall teichoic acids into the extracellular medium. J Bacteriol. 2013;195:4650–4659. doi: 10.1128/JB.00544-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Paterson GK, Tong HH, Mitchell TJ, DeMaria TF. Sortase A contributes to pneumococcal nasopharyngeal colonization in the chinchilla model. FEMS Microbiol Lett. 2005;253:151–154. doi: 10.1016/j.femsle.2005.09.052. [DOI] [PubMed] [Google Scholar]
- Comfort D, Clubb RT. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect Immun. 2004;72:2710–2722. doi: 10.1128/IAI.72.5.2710-2722.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Foster TJ. An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid. 2009;61:126–129. doi: 10.1016/j.plasmid.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Dalbey RE, Wickner W. Leader peptidase catalyzes the release of exported proteins from the outer surface of the Escherichia coli plasma membrane. J Biol Chem. 1985;260:15925–15931. [PubMed] [Google Scholar]
- Dramsi S, Trieu-Cuot P, Bierne H. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res Microbiol. 2005;156:289–297. doi: 10.1016/j.resmic.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Joyce LF, Downes J, Stockman K, Andrew JH. Comparison of five methods, including the PDM Epsilometer test (E test), for antimicrobial susceptibility testing of Pseudomonas aeruginosa. J Clin Microbiol. 1992;30:2709–2713. doi: 10.1128/jcm.30.10.2709-2713.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, Kuwano M, Heinz N, Bui NK, Hoyland CN, Ogasawara N, Lewis RJ, Vollmer W, Daniel RA, Errington J. A widespread family of bacterial cell wall assembly proteins. EMBO J. 2011;30:4931–4941. doi: 10.1038/emboj.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- Mandlik A, Swierczynski A, Das A, Ton-That H. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresso AW, Schneewind O. Sortase as a target of anti-infective therapy. Pharmacol Rev. 2008;60:128–141. doi: 10.1124/pr.107.07110. [DOI] [PubMed] [Google Scholar]
- Maresso AW, Wu R, Kern JW, Zhang R, Janik D, Missiakas DM, Duban ME, Joachimiak A, Schneewind O. Activation of inhibitors by sortase triggers irreversible modification of the active site. J Biol Chem. 2007;282:23129–23139. doi: 10.1074/jbc.M701857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci U S A. 2000;97:5510–5515. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Ton-That H, Schneewind O. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol Microbiol. 2001;40:1049–1057. doi: 10.1046/j.1365-2958.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- Mishra A, Das A, Cisar JO, Ton-That H. Sortase-Catalyzed Assembly of Distinct Heteromeric Fimbriae in Actinomyces naeslundii. J Bacteriol. 2007;189:3156–3165. doi: 10.1128/JB.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Devarajan B, Reardon ME, Dwivedi P, Krishnan V, Cisar JO, Das A, Narayana SV, Ton-That H. Two autonomous structural modules in the fimbrial shaft adhesin FimA mediate Actinomyces interactions with streptococci and host cells during oral biofilm development. Mol Microbiol. 2011;81:1205–1220. doi: 10.1111/j.1365-2958.2011.07745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Wu C, Yang J, Cisar JO, Das A, Ton-That H. The Actinomyces oris type 2 fimbrial shaft FimA mediates co-aggregation with oral streptococci, adherence to red blood cells and biofilm development. Mol Microbiol. 2010;77:841–854. doi: 10.1111/j.1365-2958.2010.07252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HV, Flores-Mireles AL, Kau AL, Kline KA, Pinkner JS, Neiers F, Normark S, Henriques-Normark B, Caparon MG, Hultgren SJ. Pilin and sortase residues critical for endocarditis- and biofilm-associated pilus biogenesis in Enterococcus faecalis. J Bacteriol. 2013;195:4484–4495. doi: 10.1128/JB.00451-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Rosini R, Rinaudo CD, Maione D, Grandi G, Telford JL. Sortase A utilizes an ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae. Infect Immun. 2008;76:3550–3560. doi: 10.1128/IAI.01613-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP. Sortase: the surface protein anchoring transpeptidase and the LPXTG motif. Trends Microbiol. 2000;8:148–151. doi: 10.1016/s0966-842x(00)01741-8. [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Nissen P. P-type ATPases. Annu Rev Biophys. 2011;40:243–266. doi: 10.1146/annurev.biophys.093008.131331. [DOI] [PubMed] [Google Scholar]
- Perry AM, Ton-That H, Mazmanian SK, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J Biol Chem. 2002;277:16241–16248. doi: 10.1074/jbc.M109194200. [DOI] [PubMed] [Google Scholar]
- Reardon-Robinson ME, Wu C, Mishra A, Chang C, Bier N, Das A, Ton-That H. Pilus hijacking by a bacterial coaggregation factor critical for oral biofilm development. Proc Natl Acad Sci U S A. 2014;111:3835–3840. doi: 10.1073/pnas.1321417111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin A, Severin A, Ritacco F, Tabei K, Singh G, Bradford PA, Siegel MM, Projan SJ, Shlaes DM. Further evidence that a cell wall precursor [C(55)-MurNAc-(peptide)-GlcNAc] serves as an acceptor in a sorting reaction. J Bacteriol. 2002;184:2141–2147. doi: 10.1128/JB.184.8.2141-2147.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F, Aebi M. Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol. 2011;21:576–582. doi: 10.1016/j.sbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Scott JR, Zahner D. Pili with strong attachments: Gram-positive bacteria do it differently. Mol Microbiol. 2006;62:320–330. doi: 10.1111/j.1365-2958.2006.05279.x. [DOI] [PubMed] [Google Scholar]
- Settem RP, Honma K, Stafford GP, Sharma A. Protein-linked glycans in periodontal bacteria: prevalence and role at the immune interface. Front Microbiol. 2014;4:310. doi: 10.3389/fmicb.2013.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpaa J, Chang C, Singh KV, Montealegre MC, Nallapareddy SR, Harvey BR, Ton-That H, Murray BE. Contribution of Individual Ebp Pilus Subunits of Enterococcus faecalis OG1RF to Pilus Biogenesis, Biofilm Formation and Urinary Tract Infection. PLoS One. 2013;8:e68813. doi: 10.1371/journal.pone.0068813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WD, Pointon JA, Abbot E, Kang HJ, Baker EN, Hirst BH, Wilson JA, Banfield MJ, Kehoe MA. Roles of minor pilin subunits Spy0125 and Spy0130 in the serotype M1 Streptococcus pyogenes strain SF370. J Bacteriol. 2010;192:4651–4659. doi: 10.1128/JB.00071-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirig T, Weiner EM, Clubb RT. Sortase enzymes in Gram-positive bacteria. Mol Microbiol. 2011;82:1044–1059. doi: 10.1111/j.1365-2958.2011.07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan A, Mandlik A, Swierczynski A, Gaspar A, Das A, Ton-That H. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol. 2007;66:961–974. doi: 10.1111/j.1365-2958.2007.05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-That H, Marraffini LA, Schneewind O. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:269–278. doi: 10.1016/j.bbamcr.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Mazmanian SK, Alksne L, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Cysteine 184 and histidine 120 of sortase form a thiolate-imidazolium ion pair for catalysis. J Biol Chem. 2002;277:7447–7452. doi: 10.1074/jbc.M109945200. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Mazmanian SK, Faull KF, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH(2)-Gly(3) substrates. J Biol Chem. 2000;275:9876–9881. doi: 10.1074/jbc.275.13.9876. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Anchor structure of staphylococcal surface proteins. IV. Inhibitors of the cell wall sorting reaction. J Biol Chem. 1999;274:24316–24320. doi: 10.1074/jbc.274.34.24316. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- Topp S, Reynoso CM, Seeliger JC, Goldlust IS, Desai SK, Murat D, Shen A, Puri AW, Komeili A, Bertozzi CR, Scott JR, Gallivan JP. Synthetic riboswitches that induce gene expression in diverse bacterial species. Appl Environ Microbiol. 2010;76:7881–7884. doi: 10.1128/AEM.01537-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roosmalen ML, Geukens N, Jongbloed JD, Tjalsma H, Dubois JY, Bron S, van Dijl JM, Anne J. Type I signal peptidases of Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:279–297. doi: 10.1016/j.bbamcr.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Wu C, Mishra A, Reardon ME, Huang IH, Counts SC, Das A, Ton-That H. Structural Determinants of Actinomyces sortase SrtC2 Required for Membrane Localization and Assembly of Type 2 Fimbriae for Interbacterial Coaggregation and Oral Biofilm Formation. J Bacteriol. 2012;194:2531–2539. doi: 10.1128/JB.00093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Mishra A, Yang J, Cisar JO, Das A, Ton-That H. Dual function of a tip fimbrillin of Actinomyces in fimbrial assembly and receptor binding. J Bacteriol. 2011;193:3197–3206. doi: 10.1128/JB.00173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Ton-That H. Allelic exchange in Actinomyces oris with mCherry fluorescence counterselection. Appl Environ Microbiol. 2010;76:5987–5989. doi: 10.1128/AEM.00811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung MK, Kozelsky CS. Transformation of Actinomyces spp. by a gram-negative broad-host-range plasmid. J Bacteriol. 1994;176:4173–4176. doi: 10.1128/jb.176.13.4173-4176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.