Abstract
Alopecia areata (AA), a cell mediated autoimmune disease, is the second most common form of hair loss in humans. While the autoimmune disease is responsible for the underlying pathogenesis, the alopecia phenotype is ultimately due to hair shaft fragility and breakage associated with structural deficits. Quantitative trait genetic analyses using the C3H/HeJ mouse AA model identified cysteine-rich secretory protein 1 (Crisp1), a hair shaft structural protein, as a candidate gene within the major AA locus. Crisp1 transcripts in the skin at various times during disease development were barely detectable. In situ hybridization identified Crisp1 expression within the medulla of hair shafts from clinically normal strains of mice but not C3H/HeJ mice with AA. Follow-up work with 5-day-old C3H/HeJ mice with normal hair also had essentially no expression of Crisp1. Other non-inflammatory based follicular dystrophy mouse models with similar hair shaft abnormalities also have little or no Crisp1 expression. Shotgun proteomics, used to determine strain difference in hair proteins, confirmed there was very little CRISP1 within normal C3H/HeJ mouse hair in comparison to 11 other strains. However, mutant mice with hair medulla defects also had undetectable levels of CRISP1 in their hair. Crisp1 null mice had normal skin, hair follicles, and hair shafts indicating that lack of the CRISP1 protein does not translate directly into defects in the hair shaft or hair follicle. These results suggest that CRISP1 may be an important structural component of mouse hair and that its strain-specific dysregulation may indicate a predisposition to hair shaft disease such as AA.
Keywords: gene array, hair shaft protein, alopecia areata predisposition
Introduction
Alopecia areata (AA), a cell mediated autoimmune disease, is the second most common form of alopecia in humans. The C3H/HeJ mouse model has been instrumental in dissecting the pathophysiology of this disease (King et al., 2008), but little is known about the cause or structural defects in the hair shafts that ultimately result in breakage (clinically evident alopecia) other than that hair keratins and melanin associated proteins have been proposed as the inciting antigens (Gilhar et al., 2001; Tobin et al., 1997). Transcriptome studies suggested that epitope spreading was a nonspecific, secondary effect of the cell mediated autoimmune aspect of AA, thereby explaining why a variety of autoantibodies were found to melanin associated and keratin proteins in both patients and controls (Carroll et al., 2002). Through studies in both mouse models and human AA patients, natural killer cell subsets are emerging as the underling effecter cells causing abnormalities in the hair follicle with secondary disruption of the normal hair shaft structure (Duncan et al., 2013; Xing et al., 2014). However, the cellular and molecular mechanisms underlying this structural damage to the hair shaft are obscure. For example, it is currently not clear whether pre-existing structural abnormalities (e.g. changes in protein composition) are involved either with the initiation of the disease process or with the severity and extent of AA.
The Hoxc13 transcription factor regulates formation of the hair shaft medulla (HSM). Dysregulation of this gene, either by overexpression in transgenic mice or lack of expression in null mice, results in defective hair shafts that break at the skin surface similar to AA. Cysteine-rich secretory protein 1 (Crisp1) expression is lost within the HSM in both of these Hoxc13 mutant mice (Peterson et al., 2005; Potter et al., 2011). Crisp1 is located on mouse chromosome 17 within the major quantitative trait locus for AA (Alaa1) (Sundberg et al., 2004). Therefore, it is possible that Crisp1 plays a role in the pathogenesis of AA in the C3H/HeJ mouse model.
This study describes the lack of CRISP1 protein and Crisp1 transcripts in the skin and hair follicles of mice that develop spontaneous AA, both mice with the disease and those that are young and clinically normal in comparison to other strains which do not develop AA. These results suggest CRISP1 is either involved in the pathogenesis predisposing to AA or its presence or absence may affect severity variability among individual patients.
Materials and methods
Mice
All procedures were done with approval by The Jackson Laboratory Animal Care and Use Committee. Mice were obtained from production colonies at The Jackson Laboratory (Bar Harbor, ME; http://jaxmice.jax.org/). The strains evaluated were AKR/J-Soat1ald/ald (strain abbreviation: AK; JR#648), BALB/cByJ (CBy; JR#1026), C3H/HeJ (C3; JR#659), C57BL/6J (B6; JR#664), CAST/EiJ (CAST; JR#928), DBA/1J (D1; JR#670), LP/J (LP; JR#676), MRL/MpJ (MRL; JR#486), MRL/MpJ-Faslpr/lpr, Foxq1sa-J/sa-J (MRL-FX; JR#3896), NOD/ShiLtJ (NOD; JR#1976), NZW/LacJ (NZW; JR#1058), RF/J (RF; JR#682), SB/LeJ-Lystbg/bg, Foxq1sa/sa (SB; JR#269), STOCK-a/a, Tmem79ma/ma, Flgft/ft/J (MAFT; JR#281), STOCK- Sgk3fz-ica/fz-ica/McirJ (FZ; JR#6135), and WSB/EiJ (WSB; JR#1145) (strain abbreviations obtained from: http://www.informatics.jax.org/external/festing/search_form.cgi; http://jaxmice.jax.org/).
Mice were maintained at The Jackson Laboratory in a humidity-, temperature-, and light cycle (12:12) controlled vivarium under specific pathogen-free conditions and were allowed free access to autoclaved food (NIH 31, 6% fat; LabDiet 5K52, Purina Mills, St. Louis, MO) and acidified water (pH 2.8–3.2).
Dorsal skin was collected from adult C57BL/6N-Crisp1tm1Pasc/tm1Pasc (1 female and 5 males), Crisp1tm1Pasc/+ (1 male and 1 female), and Crisp1+/+ (4 males) mice (Da Ros et al., 2008) at the Instituto de Biologia y Medicina Experimental (IBYME-CONICET) in Buenos Aires, Argentina. Skin was fixed in Fekete’s acid-alcohol-formalin, processed routinely, embedded in paraffin, sectioned at 6 μm, stained with hematoxylin and eosin (H&E). Hair was also collected by plucking from these same mice, mounted on glass slides with mounting media and coverslips as previously described (Silva and Sundberg, 2012). All samples were reviewed by an experienced board certified pathologist (JPS).
The microarray study was previously reported (Duncan et al., 2013; McPhee et al., 2012). Full thickness skin grafts from old C3H/HeJ mice with AA to young, histocompatible, unaffected mice were performed to induce AA in a very reproducible manner in terms of time of onset and progression (McElwee et al., 1998; Silva and Sundberg, 2013). Only female mice were used to reduce fighting, and females have a slightly higher frequency of spontaneous AA with greater clinical severity than do males (Sundberg et al., 1994a). Skin was collected and fixed by immersion in Fekete’s acid-alcohol-formalin solution for histopathology and in RNAlater (Ambion, Austin, TX) for transcriptome analyses. Skin was collected from 3 different AA graft and 3 different normal graft recipient mice at 5, 10, 15, and 20 weeks post grafting, as well as 3 different mice with spontaneous (natural) AA and 3 different normal C3H/HeJ mice.
For the in situ hybridization studies, skin was collected from 5 C3H/HeJ mice at 5 days of age and 5 at 12–18 months of age. For comparison, skin was also collected from age and gender matched FVB/NTac (Taconic, Hudson, NY).
Transcriptome analysis
Five age groups were analyzed consisting of 3 mice each with AA (15 arrays) versus age and gender matched controls (15 arrays), for a total of 30 arrays. Briefly, skin samples were stored in RNAlater (Ambion, division of Life Technologies, Grand Island, NY)). Total RNA was isolated using TRIzol and reverse transcribed with an oligo(dT)-T7 primer (Affymetrix, Santa Clara, CA), followed by double-stranded cDNA synthesisis with the Superscript (Invitrogen, division of Life Technologies, Grand Island, NY). The cDNA was amplified using T7 RNA polymerase, labeled with biotinylated nucleotides (Enzo Diagnostics, Farmingdale, NY) and was and hybridized onto the MOE430v2.0 GeneChip™ arrays (Affymetrix, Santa Clara, CA). The arrays were scanned with a GeneChip™ Scanner 3000 laser confocal slide scanner. Images were quantified using GCOS 1.0 software GeneChip™ Operating Software, Affymetrix, Santa Clara, CA). Data were transformed using RMA (Robust Multi-Array) normalization. An analysis of variance (ANOVA) model was applied to the data. Differentially expressed genes with q values < 0.05 were considered significant. Significantly different genes were then analyzed using Ingenuity Pathways Analysis® and Ariadne Genomics Pathway Studio® tools.
Shotgun Proteomics
Shotgun proteomic studies were previously reported (Rice et al., 2012) and details are available on the Mouse Phenome Database (http://phenome.jax.org/). Pelage hair from 16 mouse strains and mutant stocks were obtained by cutting hair with electric clippers from mice euthanized by CO2 asphyxiation. Hair samples were rinsed in SDS, reduced and alkylated, recovered by ethanol precipitation, and proteolyzed with stabilized bovine trypsin. The clarified digest was acidified with trifluoroacetic acid and applied to online reverse phase chromatography connected to a Thermo-Finnigan LTQ ion trap mass spectrometer. Mass spectra were extracted with Xcalibur version 2.0.7, X!Tandem was used to search the mouse Uniprot database, and Scaffold (version 3.5.1) was used to validate the peptide and protein identifications.
In situ hybridization
In situ hybridization (ISH) for Crisp1 was done as previously described (Peterson et al., 2005). For the collection of skin from 5 day postpartum, clinically normal, C3H/HeJ female and FVB/NTac mice were euthanized by decapitation. Older, 12/18 month old female C3H/HeJ mice with alopecia areata and FVB/NTac normal mice were euthanized by CO2 asphyxiation. Skin from the scapular region was fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) at 4°C overnight, followed by dehydration through a series of 5%, 15% and 30% sucrose in PBS prior to embedding in OCT compound and stored at −80°C. In situ hybridization of adult skin with Crisp1-specific digoxygenin (Roche Applied Science, Indianapolis, Indiana)-labeled RNA probes (Peterson et al. 2005) to 10 μm cryosections was performed as described (Abzhanov et al., 2003; Murtaugh et al., 1999; Pruett et al., 2004). Hybridization signals were visualized using the standard NBT/BCIP detection system (Roche).
ISH analysis of Crisp1 expression in dorsal skin of mice at 5 days postpartum was performed with a probe generated from plasmid pCrisp1-ISH. This plasmid was cloned by inserting into the EcoRI site of pSafyre vector (Bieberich et al., 1990) a central segment of 676 bp of the Crisp1 coding region that was prepared by PCR-amplification from mouse Crisp1 cDNA (GenBank: BC011150.1) using 5′CCTTGCATCATGGTCTTCTGC and 5′TGGGCTAGACTTGACTCCGA forward and reverse primer sequences, respectively.
Results and discussion
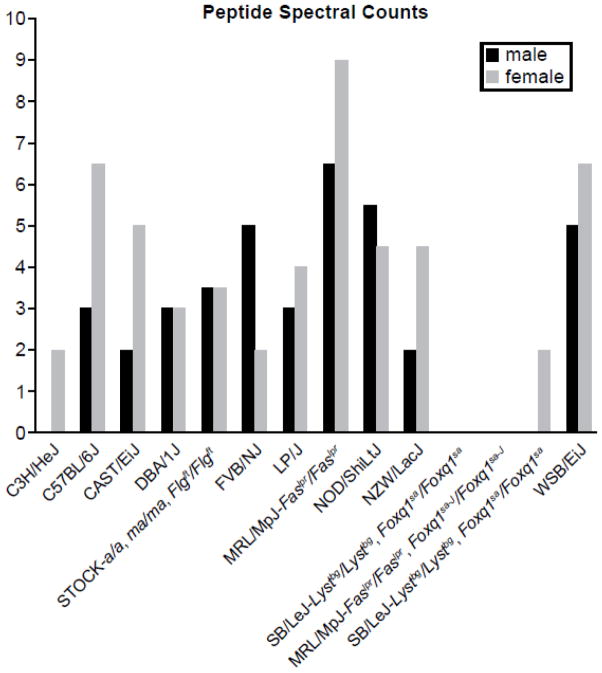
Shotgun proteomic analysis revealed low (females) to undetectable (males) levels of CRISP1 protein in normal C3H/HeJ hair shafts (Fig. 1). This finding was revealed during analysis of 11 inbred strains and 5 mutant stocks, showing that these strains were distinguishable by their protein profiles (Rice et al., 2012). CRISP1 was also undetectable in mutant mice in which the HSM did not form properly (mutations in the Foxq1 and Soat1 genes) and do not develop AA spontaneously (Wu et al., 2010; Wu et al., 2013). Targeted mutant mice, in which the Crisp1 gene had been inactivated, did not have any lesions affecting the hair follicles or hair shafts other than scattered follicular dystrophy, mild ulceration, or subepidermal fibrosis consistent with B6 alopecia and dermatitis, a common strain specific background disease in the C57BL/6 substrains (Sundberg et al., 2011).
Figure 1.
Shotgun proteomic peptide spectral counts for CRISP1protein in female and male mice from 10 inbred strains (C3H/HeJ, C57BL/6J, CAST/EiJ, DBA/1J, FVB/NJ, LP/J, MRL/MpJ- Faslpr/lpr, NOD/ShiLtJ, NZW/LacJ, and WSB/EiJ wild type mice) and inbred strains carrying filaggrin (STOCK- a/a, Tmem79ma/ma, Flgft/ft), satin (SB/LeJ-Lystbg/bg, Foxq1sa/sa), Satin-J (MRL/MpJ-Faslpr/lpr, Foxq1sa-J/sa-J, and sterol O-acyltransferase 1 (AKR/J-Soat1ald/ald) mutations. Note that C3H/HeJ males have no CRISP1 and females have very little as also seen for mutant mice with hair medulla defects (Foxq1sa, Foxq1sa-J, and Soat1).
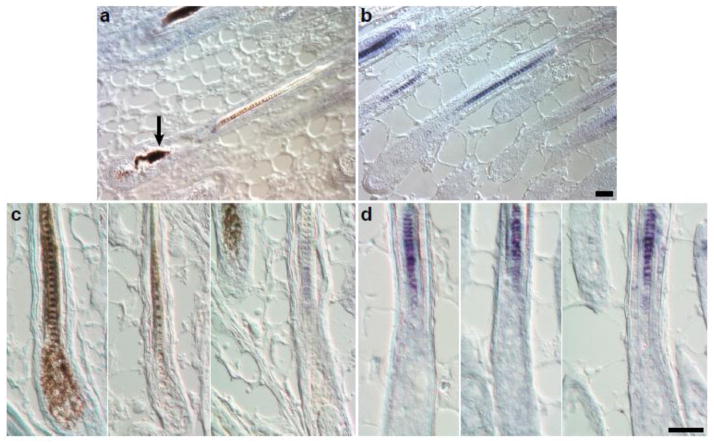
In situ hybridization to detect Crisp1 transcript expression patterns within the HSM in C3H/HeJ adult female mice with AA, due to either full thickness skin engraftment (McElwee et al., 1998) or spontaneous disease (Sundberg et al., 1994b), indicated nearly undetectable expression within the hair follicle (Fig. 2a, b). Using methods previously described (Peterson et al., 2005), no Crisp1 expression was found in C3H/HeJ adults with AA compared with normal FVB/NTac control HSM. In follow-up studies in 5 day old C3H/HeJ mice with clinically and histologically normal HSM, Crisp1 mRNA expression was also undetectable compared to age- and gender-matched FVB/NTac mouse hair where it was evident.
Figure 2. In situ hybridization for Crisp1.
There is no Crisp1 expression in representative anagen follicles from an adult AA affected C3H/HeJ mouse (A) in contrast to the blue signal in the medulla of an FVB/NTac albino mouse (B). Hair shafts were clipped from 2 adult females and 2 adult males. Note the dystrophic hair shaft in the bulb of the C3H/HeJ mouse hair follicle (A, arrow). Crisp1 expression was not detectable in normal hair shafts from 5 day old C3H/HeJ mice (C) in contrast to control 5 day old FVB/NTac mice (D). Size bar = 40μm.
Gene expression studies using both the full thickness skin graft (at 5, 10, 15, and 20 weeks after engraftment) and spontaneous disease C3H/HeJ mouse models revealed average intensity values close to background (data not shown), thereby confirming the proteomic and ISH studies.
Quantitative trait locus (QTL) analysis genetic studies of C3H/HeJ females with AA crossed with C57BL/6J mice, analyzing the F2 progeny, identified one major and three minor QTLs (Sundberg et al., 2004). Crisp1 was located within the major QTL on chromosome 17. As we showed previously, if Hoxc13, a transcription factor involved in hair medulla formation, was overexpressed (transgenic mice) or not expressed (null mutation) the hair shafts were severely dystrophic resulting in alopecia, similar to that seen in human and mouse AA. We also demonstrated that several mouse strains with single mutations in genes downstream of Hoxc13 had hair medulla defects combined with low to no expression of CRISP1 protein (Rice et al., 2012). This was further validated in adult C3H/HeJ mice with AA in which there were essentially no detectable levels of Crisp1 within the medulla. These results suggested that Crisp1 may be one of the target genes/proteins involved in the autoimmune process ultimately causing the hair shaft defect seen in AA. However, lack of Crisp1 expression was also evident in hair of 5 day old female C3H/HeJ mice as determined by ISH. This finding is consistent with data at the protein level obtained by shotgun proteomics of adult C3H/HeJ mice that had no clinical or histologic evidence of AA. Collectively, these results indicate that CRISP1 expression within the HSM of C3H/HeJ mice is greatly reduced or totally absent. Different Crisp1 alleles contain single nucleotide polymorphisms and some of these result in amino acid changes in the predicted protein sequence when compared to C57BL/6J (http://www.sanger.ac.uk/cgi-bin/modelorgs/mousegenomes/snps.pl), the normal reference strain crossed with C3H/HeJ for QTL studies (Sundberg et al., 2004), confirming these data. While C3H/HeJ mice with clinically normal hair have what appears to be a normal HSM, the medulla and entire hair shaft disintegrate when mice develop AA at specific stages of the disease leading to hair shaft breakage and alopecia. Initial studies found that male and female Crisp1 null mice exhibited no differences in fertility compared to controls, although there were functional differences in the ability of sperm to penetrate both zona pellucida-intact and zona pellucida-free eggs (Da Ros et al., 2008) suggesting that hair defects might also go unnoticed unless stressed by AA or environmental influences. These results suggest that in addition to serving as a structural component of the HSM, Crisp1 is likely to have roles that are currently unknown. Abnormalities in CRISP1 expression may predispose patients to AA or affect severity of disease.
Conclusions
While alopecia areata is a cell mediated autoimmune disorder, the ultimate cause of the clinical hair loss is follicular dystrophy with breakage of the hair shaft. This change is evident in humans and mice that develop alopecia areata. The inbred mouse strain which is prone to alopecia areata, C3H/HeJ and related substrains, have an inherent deficiency of CRISP1 protein in the hair shaft with low to undetectable levels of mRNA by gene array and in situ hybridization. These findings suggest this may predispose this strain of mice to develop alopecia or may be a complicating factor affecting severity of disease among affected individuals.
Acknowledgments
The authors thank Jesse Hammer for assistance with the figures.
This work was supported by grants from the National Institutes of Health (research grants: AR056635 and AR063781 - JPS and P42 ES04699 - RHR; core services: CA034196 - The Jackson Laboratory). The National Alopecia Areata Foundation (NAAF) helped to support the mouse model development.
Abbreviations
- AA
alopecia areata
- Alaa1
alopecia areata quantitative trait locus 1
- Crisp1/CRISP1
cysteine-rich secretory protein 1, gene/PROTEIN
- Crisp1tm1Pasc
Crisp1 targeted (knockout) mutant mouse line
- Foxq1
forkhead box q1 gene
- HSM
hair shaft medulla
- Hoxc13
homeobox C13 gene
- ISH
in situ hybridization
- QTL
quantitative trait locus
- Soat1
sterol-acyltransferase 1, gene
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abzhanov A, et al. Dissimilar regulation of cell differentiation in mesencephalic (cranial) and sacral (trunk) neural crest cells in vitro. Development. 2003;130:4567–4579. doi: 10.1242/dev.00673. [DOI] [PubMed] [Google Scholar]
- Bieberich CJ, et al. Evidence for positive and negative regulation of the Hox-3.1 gene. Proc Natl Acad Sci USA. 1990;87:8462–8466. doi: 10.1073/pnas.87.21.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J, et al. Gene array profiling and immunomodulation studies define a cell mediated immune response underlying the pathogenesis of alopecia areata in a mouse model and humans. J Invest Dermatol. 2002;119:392–402. doi: 10.1046/j.1523-1747.2002.01811.x. [DOI] [PubMed] [Google Scholar]
- Da Ros VG, et al. Impaired sperm fertilizing ability in mice lacking Cysteine-RIch Secretory Protein 1 (CRISP1) Develop Biol. 2008;320:12–18. doi: 10.1016/j.ydbio.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan FJ, et al. Endogenous retinoids in the pathogenesis of alopecia areata. J Invest Dermatol. 2013;133:334–343. doi: 10.1038/jid.2012.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilhar A, et al. Melanocyte-associated T cell epitopes can function as autoantigens for transfer of alopecia areata to human scalp explants on Prkdcscid mice. J Invest Dermatol. 2001;117:1357–1362. doi: 10.1046/j.0022-202x.2001.01583.x. [DOI] [PubMed] [Google Scholar]
- King LE, et al. Alopecia areata. In: Nickoloff BJ, Nestle FO, editors. Dermatologic Immunity: Current Directions in Autoimmunity. Karger; Basel: 2008. pp. 280–312. [DOI] [PubMed] [Google Scholar]
- McElwee KJ, et al. Experimental induction of alopecia areata-like hair loss in C3H/HeJ mice using full-thickness skin grafts. J Invest Dermatol. 1998;111:797–803. doi: 10.1046/j.1523-1747.1998.00380.x. [DOI] [PubMed] [Google Scholar]
- McPhee CG, et al. Gene expression studies identify Cxcr3 and its ligands, Cxcl9, Cxcl10, and Cxcl11 in the pathogenesis of alopecia areata in the mouse. J Invest Dermatol. 2012;132:1736–1738. doi: 10.1038/jid.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC, et al. Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev. 1999;13:225–237. doi: 10.1101/gad.13.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RL, et al. Epididymal cysteine-rich secretory protein 1 (CRISP-1) encoding gene is expressed in murine hair follicles and downregulated in mice overexpressing Hoxc13. J Investig Dermatol Symp Proc. 2005;10:238–242. doi: 10.1111/j.1087-0024.2005.10114.x. [DOI] [PubMed] [Google Scholar]
- Potter CS, et al. The nude mutant gene Foxn1 is a HOXC13 regulatory target during hair follicle and nail differentiation. J Invest Dermatol. 2011;131:828–837. doi: 10.1038/jid.2010.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruett ND, et al. Krtap16, characterization of a new hair keratin-associated protein (KAP) gene complex on mouse chromosome 16 and evidence for regulation by Hoxc13. J Biol Chem. 2004;279:51524–51533. doi: 10.1074/jbc.M404331200. [DOI] [PubMed] [Google Scholar]
- Rice RH, et al. Differentiating inbred mouse strains from each other and those with single gene mutations using hair proteomics. PLos One. 2012;7:e51956. doi: 10.1371/journal.pone.0051956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva KA, Sundberg JP. Necropsy methods. In: Hedrich HJ, editor. The laboratory mouse. Academic Press; London: 2012. pp. 779–806. [Google Scholar]
- Silva KA, Sundberg JP. Surgical methods for full thickness skin grafts to induce alopecia areata in C3H/HeJ mice. Comp Med. 2013;63:392–397. [PMC free article] [PubMed] [Google Scholar]
- Sundberg JP, et al. Alopecia areata in aging C3H/HeJ mice. J Invest Dermatol. 1994a;102:847–856. doi: 10.1111/1523-1747.ep12382416. [DOI] [PubMed] [Google Scholar]
- Sundberg JP, et al. Adult onset alopecia areata is a complex polygenic trait in the C3H/HeJ mouse model. J Invest Dermatol. 2004;123:294–297. doi: 10.1111/j.0022-202X.2004.23222.x. [DOI] [PubMed] [Google Scholar]
- Sundberg JP, et al. Primary follicular dystrophy with scarring dermatitis in C57BL/6 mouse substrains resembles central centrifugal cicatricial alopecia in humans. Vet Pathol. 2011;42:513–524. doi: 10.1177/0300985810379431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg JP, et al. Alopecia areata in aging C3H/HeJ mice. In: Sundberg JP, editor. Handbook of mouse mutations with skin and hair abnormalities: animal models and biomedical tools. CRC Press; Boca Raton: 1994b. pp. 499–505. [Google Scholar]
- Tobin DJ, et al. Autoantibodies to hair follicles in C3H/HeJ mice with alopecia areata-like hair loss. J Invest Dermatol. 1997;109:329–333. doi: 10.1111/1523-1747.ep12335848. [DOI] [PubMed] [Google Scholar]
- Wu B, et al. Mutations in sterol O-acyltransferase 1 (Soat1) results in hair interior defects in AKR/J mice. J Invest Dermatol. 2010;130:2666–2668. doi: 10.1038/jid.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, et al. R164C mutation in FOXQ1 H3 domain affects formation of the hair medulla. Exp Dermatol. 2013;22:234–236. doi: 10.1111/exd.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, et al. Alopecia areata is driven by cytotoxic T lymphocytes and is reversed by JAK inhibition. Nat Med. 2014;20:1043–1049. doi: 10.1038/nm.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]