Figure 1.
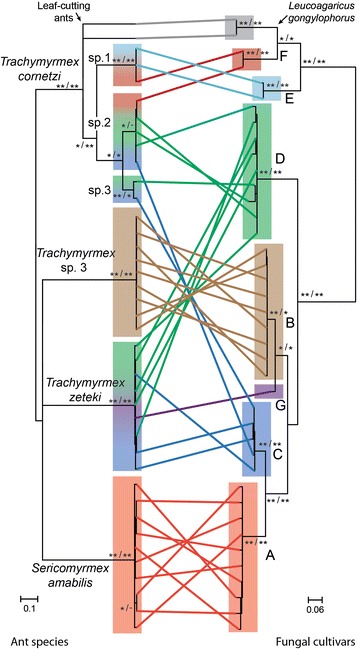
Interaction specificity between Panamanian higher-attine ants and cultivars. Fungus-growing ant and cultivar co-phylogeny color coded for each colony based on network analyses that independently identified groupings corresponding to seven fungal symbiont ITS haplotypes (A-G) and six ant species (S. amabilis, T. zeteki, T. sp. 3, and T. cornetzi sp. 1–3). T. zeteki and three T. cornetzi species (two of them cryptic and new to science) share a variable group of fungal ITS haplotypes (C-G), whereas S. amabilis and T. sp. 3 each appear to cultivate a single ITS haplotype (A red and B brown, respectively). The phylogenies are based on maximum-likelihood and Bayesian analysis of a 549 bp alignment of partial CO1, tRNA, and CO2 ant sequences and a 740 bp alignment of ITS1, 5.8S, and ITS2 fungal sequences. The analyses included ant and cultivar sequences for the leaf-cutting ant species Acromyrmex octospinosus and Atta cephalotes (grey), which as expected from recent phylogenetic analyses branched off closer to the T. cornetzi lineage than the Sericomyrmex/Trachymyrmex zeteki clade [25]. Both trees are mid-point rooted and branch support is shown as: Maximum-likelihood (100 = **, 95 < *)/Bayesian (1.00 = **, 0.95 < *). Scale bars are substitutions per site.
