Figure 2.
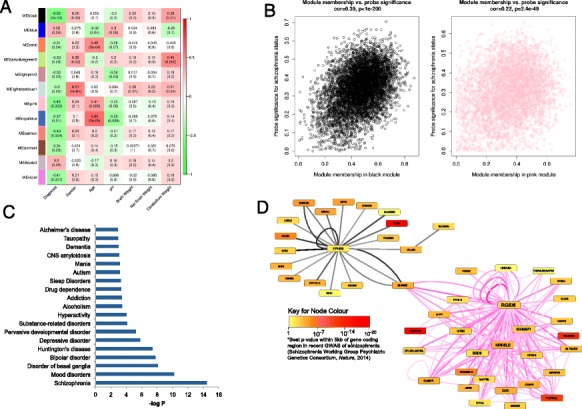
Schizophrenia-associated gene co-methylation modules in the prefrontal cortex. (A) Heatmap representing correlations between module eigenvalues (ME) and clinical/demographic variables. Each row represents a specific module, indicated by an arbitrary module color labeled on the y-axis. Dark red indicates strong positive correlation, dark green indicates strong negative correlation, and white indicates no correlation, as indicated in color scale bar. P-values are given in parentheses. The black (P = 0.0004) and pink (P = 0.002) WGCNA modules are most significantly associated with schizophrenia in the PFC. (B) Module membership in both modules is strongly correlated with probe significance with disease (black module: r = 0.39, P <1e-200; pink module: r = 0.22, P = 2.4e-49). (C) Ingenuity Pathway Analysis of genes associated with CpG sites in the black module reveals a highly significant enrichment of disease pathways related to schizophrenia and other neuropsychiatric disease. (D) Co-methylation networks between the top 1% of loci in the black and pink modules ranked by their module membership, where thicker lines indicate more connected genes. Black edges represent co-methylation between probes in the black module and pink edges represent co-methylation in the pink module; thicker and deeper colored edges indicate stronger correlations between probes. Interestingly, the two modules are connected by SHANK2, which encodes a molecular scaffold protein that plays a role in synaptogenesis. Furthermore, several of the genes are associated with schizophrenia in recent collaborative genome-wide association studies (GWAS) [27]. GWAS significance for SNPs in the vicinity of each locus is depicted by color.
