Figure 4.
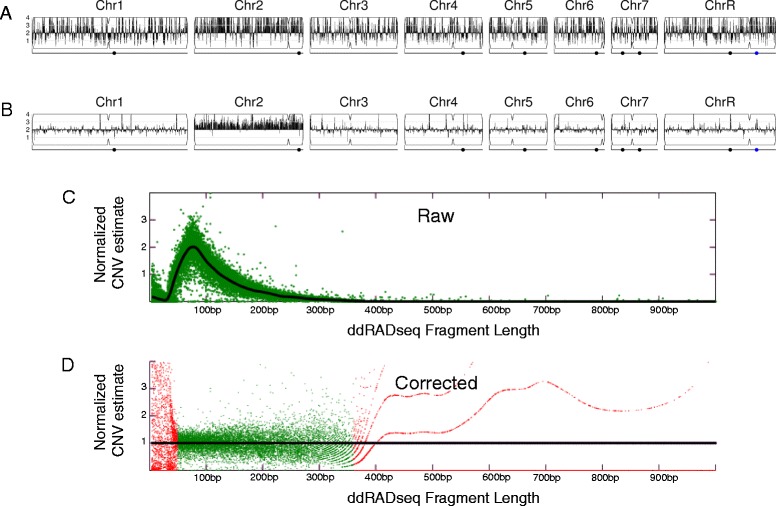
Normalization of fragment-length-bias in ddRADseq data. (A) High noise of raw read-depth CNV estimates in CHY477 [35] ddRADseq data with GC-content, fragment-length, and position-effect biases. (B) CNV estimates mapped across the genome and corrected for GC bias, fragment length bias and normalized to the reference data. (C) Average read-depth CNV estimates versus predicted restriction fragment length for strain RBY917 Mata/a -his, -leu, delta gal1::SAT1/GAL1 derived from SNY87 [36]. Black, LOWESS fit curve. (D) Corrected average read-depth CNV estimates versus fragment length, with regions of low reliability data in red, as described in more detail in the text. Chromosome illustrations are as in Figure 2.
