Figure 7.
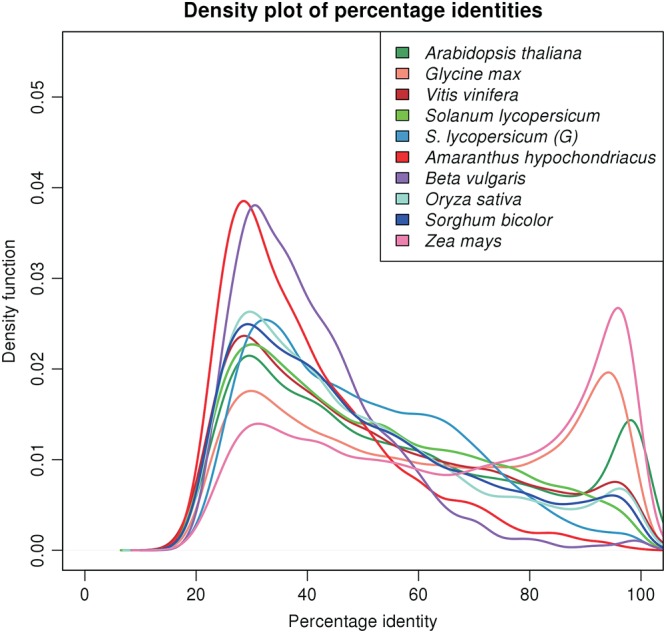
A genome duplication study in A. thaliana, G. max, V. vinifera, S. lycopersicum, A. hypochondriacus, B. vulgaris, O. sativa, S. bicolor, and Z. mays by comparison of the distribution of percentage identities among paralogous pairs from the respective proteomes. The GENSCAN-predicted proteome of S. lycopersicum is labelled as S. lycopersicum (G) in the figure. The Kernel density plot of the density function of the number of paralogous pairs against the percentage identities for the diverse sequenced plant species shows the first peak at 20–40% identity corresponding to the paleaohexaploidy event and the sharp peak at 90–100% identity representing a recent whole genome duplication event. From the plot, it is evident that A. hypochondriacus, like B. vulgaris, did not undergo any recent whole genome duplication.
