Figure 2.
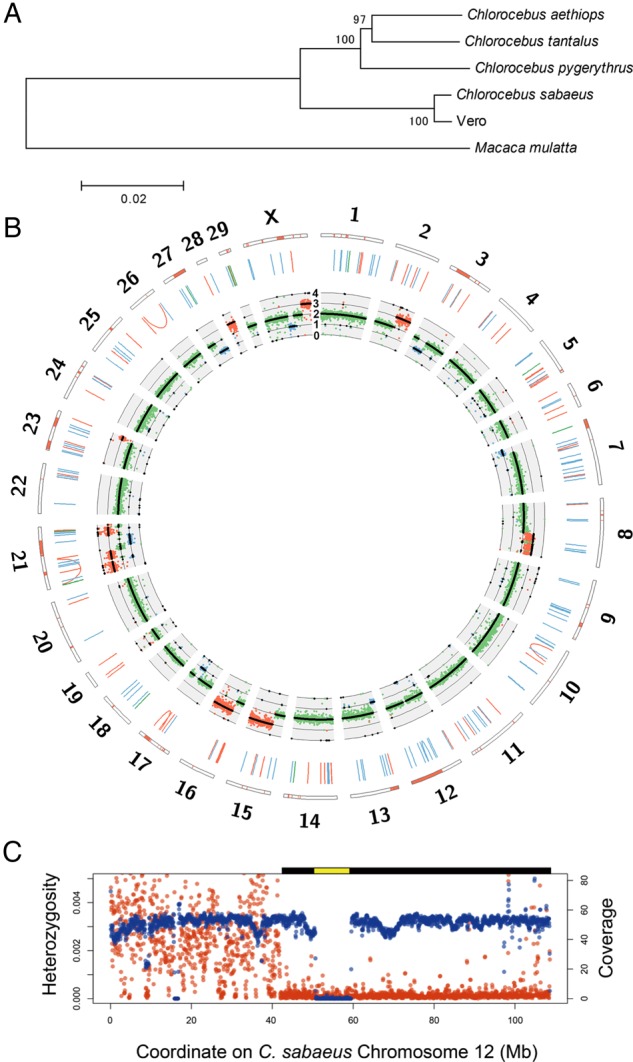
Genome landscape of the Vero genome. (A) Phylogeny of mitochondrial genomes of the Vero cell line, four Chlorocebus species, and Macaca mulatta. Bootstrap values with 1,000 replications were shown upon the branches. (B) Circos plot of the Vero cell genome. The orange bars in the outermost rectangles represent LOH regions. The blue, green, and orange lines in the middle layer show deletions, duplications, and inversions larger than 1 kb, respectively. The innermost plot shows the coverage of paired-end reads and expected ploidy (black lines). The blue, green, and orange dots represent the coverage values in 1×, 2×, and 3× regions, respectively. (C) The large deletion and LOH regions on Chlorocebus sabaeus chromosome 12. The red and blue points represent average heterozygosity (the number of heterozygous SNVs per bp) and genome coverage of paired-end reads in 1-Mb-size windows, respectively. The predicted homozygous deletion regions and LOH regions are shown as yellow and black bars on the plot area, respectively.
