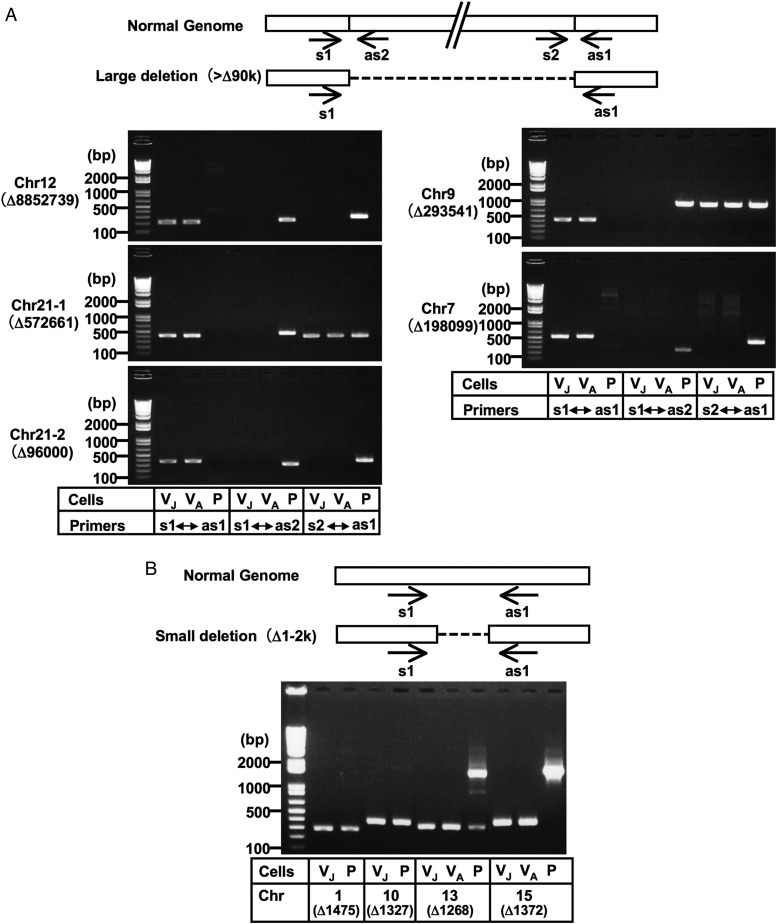
Figure 3.
Validation of genomic deletions in Vero cells by PCR analysis. Large deletions (>90 kb deletions) (A) and some small deletions (1–2 kb deletions) (B) were selected. Although the genomic PCR confirmed the existence of breakpoint junctions for the 573 kb deletion in chromosome 21 and the 294 kb deletion in chromosome 9, a part of these regions appeared to exist somewhere in the genome (A; see also Supplementary Fig. S2). Amplicons corresponding to the deletions predicted in chromosomes 1 and 10 were produced not only from Vero cells, but also from AGM PBMC, while amplicons corresponding to the ‘non-deleted’ counterparts were not produced even from AGM PBMC (B; see also Supplementary Fig. S2), which indicated that our determined sequences for the Vero cell genome existed homozygously in these regions not only in Vero cells, but also in AGM PBMC. This paradox might be attributed to the possible incompleteness of the currently available version of the AGM whole-genome draft sequence or polymorphic state of the deletion within AGM populations. Normal Genome indicates the sequences predicted from the draft genome sequences of AGM and the rhesus macaque. Arrows indicate the primer positions used in the PCR analyses. The ‘Δ’ indicates the genomic deletion size predicted by the massively parallel sequencing system. The templates used were as follows: VJ, Vero JCRB0111; VA, Vero ATCC; P, AGM PBMC. PCR amplicons were sequenced to confirm the breakpoint sequences, which are shown in Supplementary Fig. S2. Chr, chromosome.