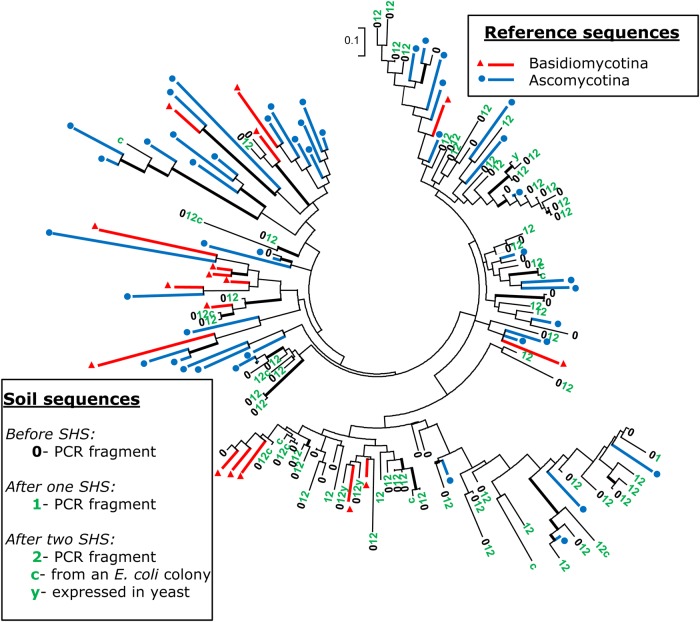
Figure 4.
Phylogenetic diversity of the GH11 partial amino acid sequences obtained from PUE cDNA samples. 0, 1 and 2 translated PCR sequences obtained before or after one or two cycles of hybridization. PUE sequences are scattered over the entire tree that includes representative reference sequences from Ascomycota and Basidiomycota. c, sequences obtained from Escherichia coli clones; y, sequences functionally expressed in yeast clones. PhyML tree calculation was based on an alignment of ca. 80-amino-acid-long GH11 partial sequences. Thicker internal black branches indicate bootstrap value ≥60% (1,000 replications). Full species names and accession numbers of the reference sequences are given in Supplementary Fig. S6A. Similar trees drawn using the sequences from sites BRE, BRH and BEW are illustrated in Supplementary Fig. S6 B, C and D, respectively. This figure appears in colour in the online version of DNA Research.