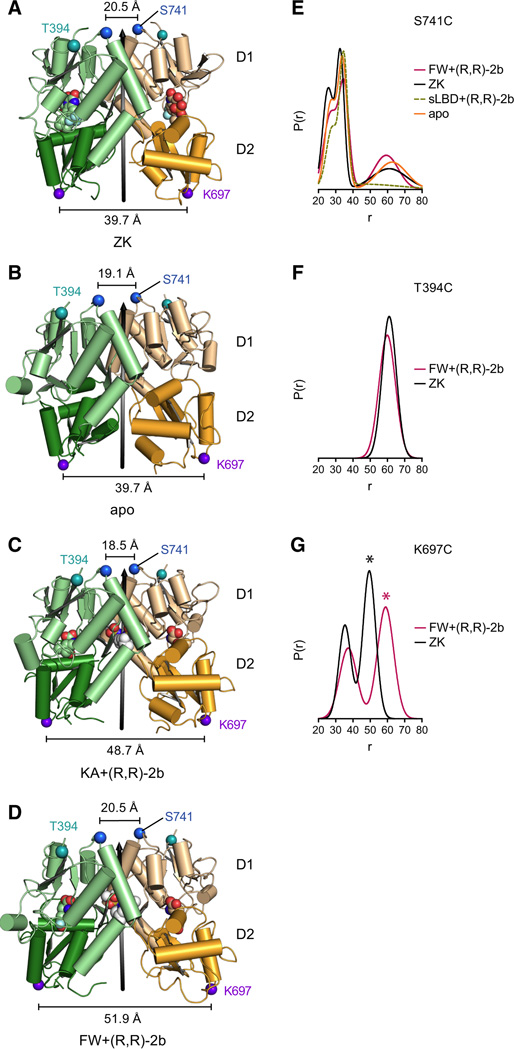
Figure 3. Intra-dimer LBD interfaces in apo and partial agonist-bound states.
A–D Side views of LBD dimers from chain A (green) and chain D (yellow) of full-length structures in complex with the competitive antagonist ZK (A; PDB code: 3KG2), in the apo state (B), in complex with partial agonist KA + modulator (R,R)-2b (C) and in complex with partial agonist FW + modulator (R,R)-2b (D). D1 domains are colored in lighter shades. LBD local 2-fold axes are shown as black arrows. Distances between Cα atoms (blue spheres) of residues Ser 741 are indicated by black scale bars. Distances between Cα atoms (purple spheres) of residues Lys 697 are indicated by black scale bars. Cα atoms of marker position T394C are shown as teal spheres. Modulator (R,R)-2b is shown in white space-filling representation. E-G. Probability distributions of DEER distances calculated from DEER decays, provided in Figure S4 E–G, of MTSSL-labeled receptor constructs S741C (E), T394C (F) and K697C (panel G) measured under apo conditions (orange), with ZK (black), or with FW+(R,R)-2b (magenta), respectively. Note that the long-distance peak of ~60 Å present under all conditions in the DEER distribution of mutant S741C (in F) is dependent on the background correction. The distance distribution of the respective MTSSL-labeled sLBD S741C construct in presence of FW+(R,R)-2b is superposed as yellow dashed line (E). The asterisk in panel G indicates putative inter-dimer distances.