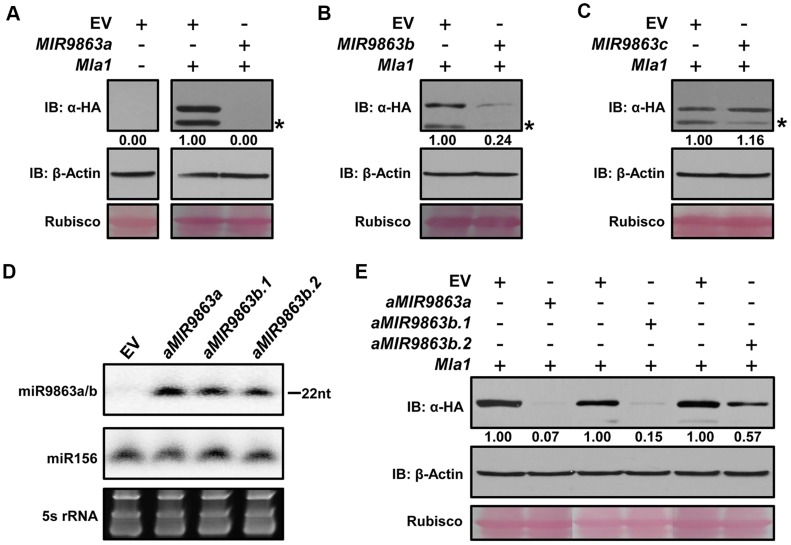
Figure 2. miR9863a, miR9863b.1 and miR9863b.2, but not miR9863c, regulate Mla1.
(A) to (C) miR9863 family members differentially regulate Mla1 gene shown by transient gene expression assay in N. benthamiana. Agrobacteria strain expressing Mla1-3HA were co-infiltrated into N. benthamiana leaves with strains containing the respective miRNA precursors, hvu-MIR9863a (A), hvu-MIR9863b (B) or tae-MIR9863c (C). Western blotting was used to detect MLA1-3HA or actin levels at 36 hours post Agro-infiltration (hpai); and the number below the blots indicates relative protein levels as calculated by Image J. Empty vector (EV) was used as a negative control and rubisco served as a loading control. The asterisks indicate non-specific signals. Experiments were performed at least two times with similar results. (D) Expression of mature miR9863a, miR9863b.1 and miR9863b.2 using artificial MIRNA backbone. The mature miR9863a, miR9863b.1 or miR9863b.2 sequences were engineered into the Arabidopsis MIR173 precursor (see S5 Figure) and then artificial MIRNA precusors (aMIRNA) were expressed in N. benthamiana by Agro-infiltration. The levels of mature miR9863s were detected by RNA gel blot analysis at 36 hpai. miR156 and 5S rRNA were employed as loading controls. (E) Determination of the regulation efficiency of miR9863a, miR9863b.1 or miR9863b.2 on Mla1. The aMIR9863a, aMIR9863b.1 or aMIR9863b.2 was respectively co-expressed with Mla1 in N. benthamiana, and MLA1 protein levels were determined at 36 hpai by immunoblotting.