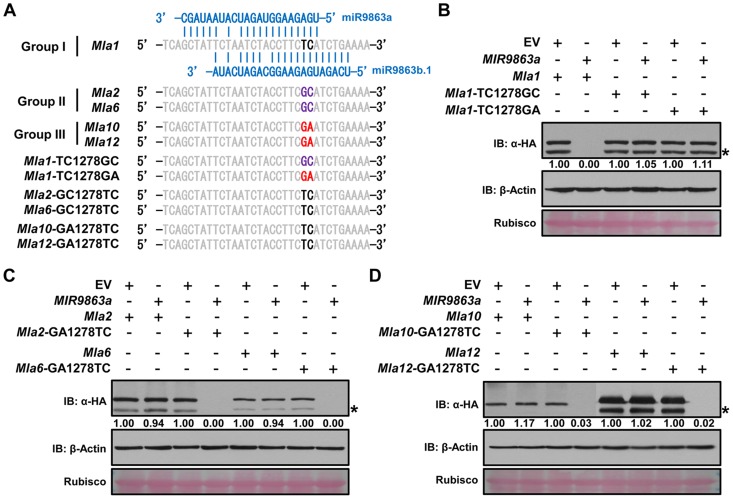
Figure 4. SNP variations in Mla alleles dictate miR9863 specificity.
(A) Alignment of miR9863 target site of WT and mutated Mla alleles. SNPs in bold font and color-coded represent Mla type of group I (black), II (magenta) or III (red). Nucleotide substitutions TC1278GC or TC1278GA were introduced into Mla1; GC1278TC into Mla2 and Mla6, and GA1278TC into Mla10 and Mla12. (B) to (D) Determination of miR9863a regulation specificity on WT or mutated Mla alleles from different groups. Using transient gene expression assays in N. benthamiana, Hvu-MIR9863a was respectively co-expressed with group I Mla1 alleles (B), or group II Mla2 and Mla6 alleles (C), or group III Mla10 and Mla12 alleles (D). The protein accumulation levels of indicated Mla isoforms and actin were determined by immunoblotting. The asterisks indicate non-specific signals.