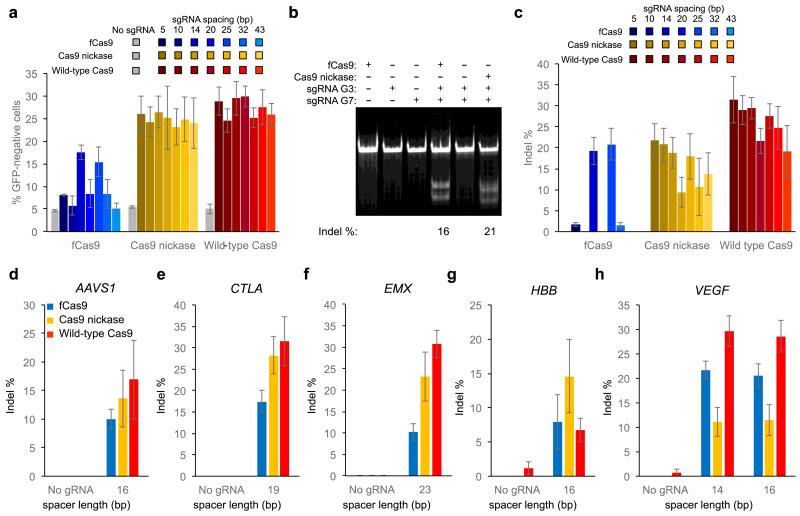
Figure 2. Genomic DNA modification by fCas9, Cas9 nickase, and wild-type Cas9.
Detection of genomic modification by loss of GFP signal or Surveyor assay at either an integrated GFP gene, or at endogenous genomic targets within the AAVS1, CLTA, EMX, HBB, or VEGF genes (Supplementary Figure 5) (a) GFP disruption activity of fCas9, Cas9 nickase, or wild-type Cas9 with either no sgRNA, or sgRNA pairs of variable spacer length targeting the GFP gene in orientation A. (b) Indel modification efficiency from PAGE analysis of a Surveyor cleavage assay of renatured target-site DNA amplified from cells treated with fCas9, Cas9 nickase, or wild-type Cas9 and two sgRNAs spaced 14 bp apart targeting the GFP site (sgRNAs G3 and G7; Figure 1d), each sgRNA individually, or no sgRNAs. The indel modification percentage is shown below each lane for samples with modification above the detection limit (~2%). (c-h) Indel modification efficiency for (c) two pairs of sgRNAs spaced 14 or 25 bp apart targeting the GFP site, (d) on pair of sgRNAs spaced 16 bp apart targeting the AAVS1 site, (e) one pair of sgRNAs spaced 19 bp apart targeting the CLTA site, (f) one pair of sgRNAs spaced 23 bp apart targeting the EMX site (g) one pair of sgRNAs spaced 16 bp apart targeting the HBB site, and (h) two pairs of sgRNAs spaced 14 or 16 bp apart targeting the VEGF site. Error bars reflect standard error of the mean from three biological replicates performed on different days.