Figure 6.
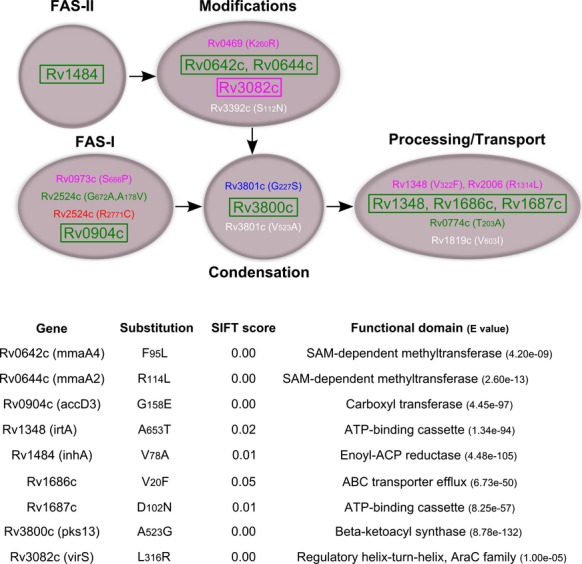
Lineage-specific nonsynonymous single-nucleotide polymorphisms (nsSNPs) from “ancient” lineages are predicted deleterious. nsSNPs specific to each Mycobacterium tuberculosis complex (MTBC) lineage were extracted and the related genes listed and grouped according to their involvement in the different step of the metabolic pathway of mycolic acid biosynthesis (white color code was used for SNPs shared by the two “modern” lineages). The amino acid substitutions were subjected to SIFT algorithm to predict loss of function based on the degree of conservation of amino acid residues in sequence alignments derived from closely related sequences (Sim et al. 2012). Genes highlighted in boxes presented substitutions with a statistically significant probability to affect protein function (P < 0.05) whereas others were predicted to be tolerated. Gene names, amino acid substitution, SIFT score as well as the predicted domain affected by the mutation are presented for deleterious substitutions.
