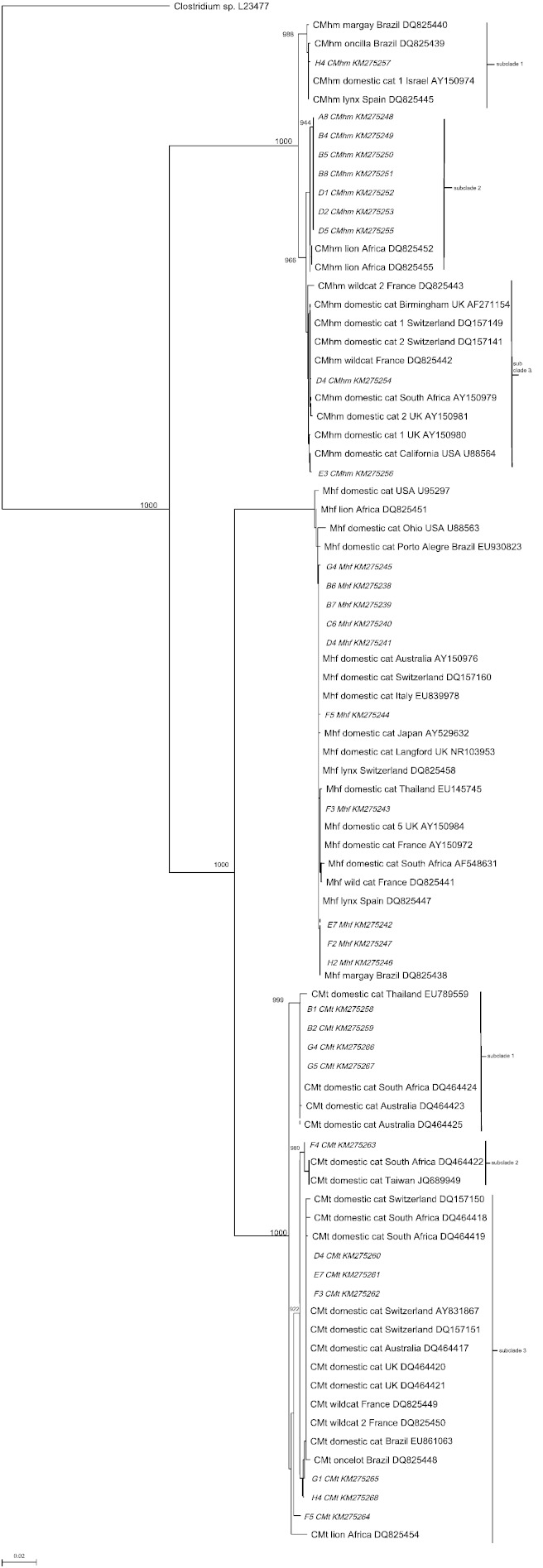
Fig. 2.
Phylogenetic analysis of near complete 16S rRNA gene sequences from Mhf, CMhm and CMt species in cats from Brasília and surrounding areas. The Neighbor-joining method was used to construct the phylogenetic tree with bootstrap values provided at the nodes (only values ≥ 700 are shown). Evolutionary distances are to the scales shown. Clostridium spp. was used as an out-group. GenBank accession numbers are indicated in the figure. Mhf sequences (B6, B7, C6, D4, E7, F2, F3, F5, G4, H2), CMhm sequences (A8, B4, B5, B8, D1, D2, D4, D5, E3, H4) and CMt sequences (B1, B2, D4, E7, F3, F4, F5, G1, G4, G5, H4) were generated in the current study. Mhf = Mycoplasma haemofelis, CMhm = “Candidatus Mycoplasma haemominutum”, CMt = “Candidatus Mycoplasma turicensis”.