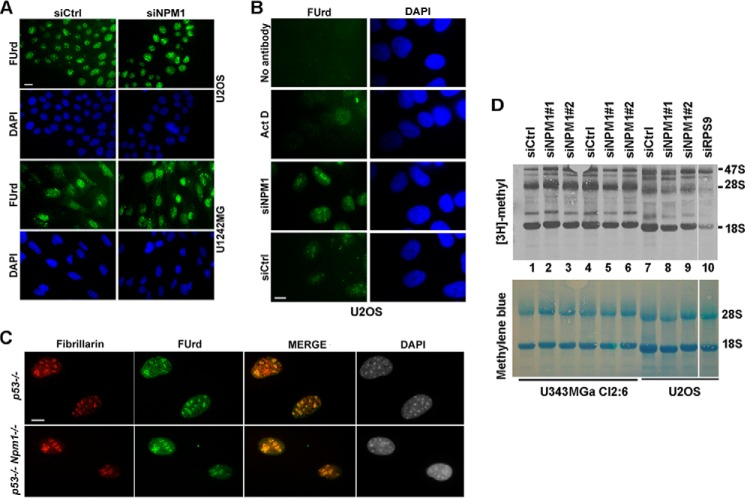
FIGURE 10.
rDNA transcription and processing in NPM1-depleted cells. A, 5-FUrd incorporation in NPM1-depleted U1242MG and U2OS cells. The cells were incubated in 2 mm 5-FUrd for 10 min. Scale bar: 20 μm. B, control reactions for 5-FUrd labeling included no primary antibody control and 5-FUrd labeling in actinomycin D-treated U2OS cells (10 nm actinomycin D (Act D) for 2 h) resulting in a much reduced signal. As a comparison, 5-FUrd labeling is also shown in siNPM1- and siCtrl-treated U2OS cells. Scale bar: 10 μm. C, 5-FUrd incorporation in p53−/− and p53−/−,Npm1−/− cells. The cells were incubated in 2 mm 5-FUrd for 10 min. Incorporation of 5-FUrd into newly made RNA in A–C was detected by a primary antibody against BrdU followed by a secondary FITC-conjugated anti-mouse antibody. In C, cells were also stained for fibrillarin, revealing co-localization of 5-FUrd and fibrillarin in nucleoli. Scale bar: 10 μm. D, synthesis and processing of rRNA. U2OS cells (lanes 7–10) or U343MGa Cl2:6 cells (lanes 1–6) were transfected with siRNA oligonucleotides as indicated. Lanes 4–6 are a replicate experiment of lanes 1–3. Knockdown and control cells were labeled with [methyl-3H]methionine for 2 h. Ribosomal protein S9 (RPS9) siRNA served as a positive control for disrupted processing of rRNA (31). Newly synthesized 47S, 28S, and 18S rRNA species are indicated (upper panel), and the total 28S and 18S rRNA transferred to the membrane was visualized by methylene blue (lower panel).