Figure 1.
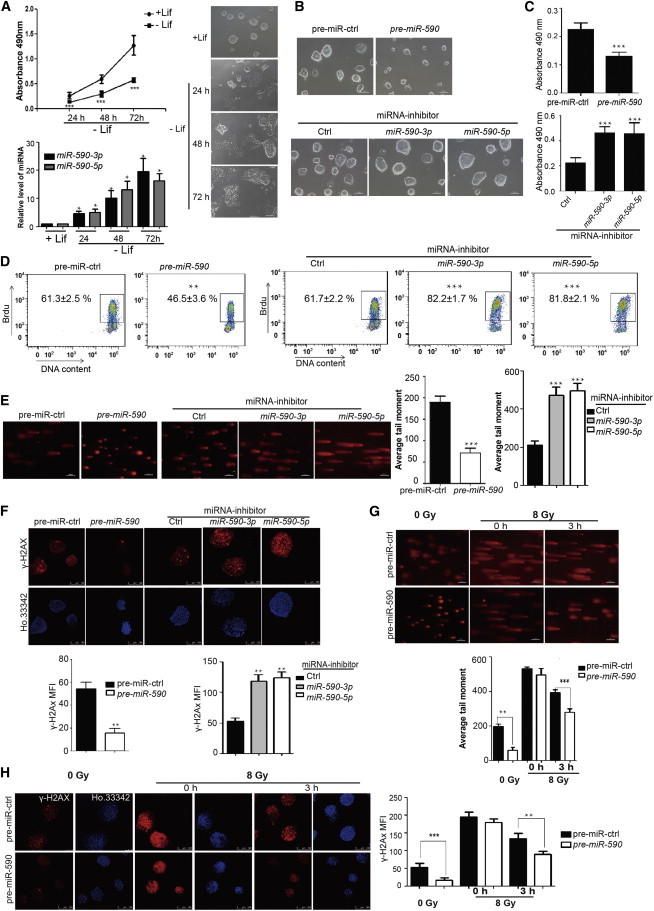
miR-590 Inhibits Rapid Proliferation and Promotes SSB and DSB Damage Repair of mESCs
(A) Analysis of the proliferation of mESCs cultured with or without LIF by MTS cell proliferation assay. (Bottom) The qRT-PCR verification of miR-590-3p and miR-590-5p levels. (Right) The morphology of mESCs cultured with (+LIF) on 24 hr or without LIF (−LIF) on 24, 48, 72 hr. Data shown are means ± SD of six independent experiments (n = 6). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. The scale bar represents 100 μm.
(B) Morphology of colonies of mESCs after transfection with pre-miR-590 or miR-590-3p/5p inhibitor. pre-miR-ctrl indicates pre-miRNA control, which has no homology to any known mammalian gene. Inhibitor control is a random sequence molecule that has no homology to any known mammalian gene. The scale bar represents 100 μm.
(C) Analysis of the proliferation of mESCs transfected with pre-miR-590 or miR-590-3p/5p inhibitor by MTS cell proliferation assay. Data shown are means ± SD of five independent experiments (n = 5). ∗∗∗p < 0.001.
(D) Analysis of the proliferation of mESCs transfected with pre-miR-590 or miR-590-3p/5p inhibitor by FACS analysis of BrdU incorporation. The figure shows the percentage (%) of population the of BrdU-positive cells. Data shown are means ± SD of three independent experiments (n = 3). ∗∗p < 0.01 and ∗∗∗p < 0.001.
(E) Comet assay for SSB damage in mESCs. The scale bar represents 100 μm. Data shown are means ± SD of three independent experiments (n = 3). (Right) The quantification of the average DNA tail moment. ∗∗∗p < 0.001.
(F) Immunofluorescence analysis of γ-H2AX (red) to indicate the state of DSB damage in mESCs. Ho.33342 (Hoechst 33342) represents nuclear staining (blue). The scale bar represents 100 μm. (Bottom) The quantification of the fluorescence of γ-H2AX immunostaining. Data shown are means ± SD of three independent experiments (n = 3). ∗∗p < 0.01. MFI, mean fluorescence intensity.
(G) Comet assay for SSB damage of mESCs with irradiation from X-ray. 0 Gy indicates mESCs not subjected to X-ray irradiation. The scale bar represents 100 μm. The bottom shows the quantification of the average DNA tail moment. Data shown are means ± SD of three independent experiments (n = 3). ∗∗p < 0.01 and ∗∗∗p < 0.001.
(H) Immunofluorescence analysis of γ-H2AX (red) with Ho.33342 (Hoechst 33342) nuclear staining (blue). The scale bar represents 100 μm. (Right) The quantification of the fluorescence of γ-H2AX immunostaining. Data shown are means ± SD of three independent experiments (n = 3). ∗∗p < 0.01 and ∗∗∗p < 0.001.
