Figure 2.
Transcriptional and Epigenetic Reprogramming of iNSCs
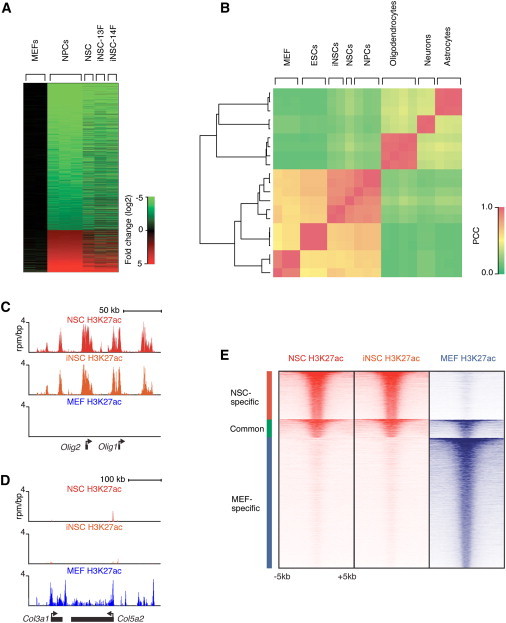
(A) Global gene expression analysis comparing the untransduced MEF starting population (P3), primary-derived control NSCs, iNSC-13F, and iNSC-14F (all P8). The heatmap indicates the fold change (log2) of gene expression relative to MEFs. Differentially expressed genes were determined by comparing published ESC-derived NPC and MEF data sets and arranged in rows (Mikkelsen et al., 2008).
(B) Hierarchical clustering and Pearson correlation analysis of gene expression in iNSCs (iNSC-13F and iNSC-14F), control NSCs, ESCs, ESC-derived NPCs, differentiated neural cell lineages, and MEFs (Mikkelsen et al., 2008; Cahoy et al., 2008). The color indicates the Pearson correlation coefficient (PCC).
(C and D) H3K27ac ChIP-seq profiles of the Olig1/Olig2 locus (C) or the Col3a1/Col5a2 locus (D) in control NSCs, iNSC-13F (iNSC) (both P8), and MEFs (P3).
(E) Heat map representation of enhancer-associated histone modification H3K27ac at the union of 16,706 enhancer regions from NSCs and MEFs. The enhancer regions were grouped into NSC-specific, common, and MEF-specific enhancers. Read density surrounds the center (±5 kb) of enhancer regions, rank ordered from highest to lowest average H3K27ac occupancy within each group.
See also Figure S2.
