Table 1.
Identification of 21 proteins that differentially expressed in labellum and inner lateral petal.
| Group a | Spot Number b | Protein Name | Reference Organism | Accession c | Mascot Scores | Blast Score | Blast Expect | Spots % Volume Variations (p < 0.05) d |
|---|---|---|---|---|---|---|---|---|
| I | 173 | Superoxide dismutase [Cu-Zn], chloroplastic | Oryza sativa japonica group | P93407 | 214 | 306 | 9 × 10−103 | 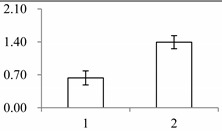 |
| I | 243 | 2-Cys peroxiredoxin BAS1, chloroplastic | Arabidopsis thaliana | Q96291 | 51 | 376 | 6 × 10−129 | 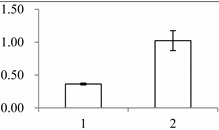 |
| I | 302 | Triosephosphate isomerase (TPI) | Gossypium mexicanum | D2D303 | 137 | 424 | 2 × 10−147 | 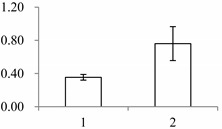 |
| I | 411 | Fibrillin-like protein (FIB) | Oncidium hybrid cultivar | B4F6G1 | 930 | 489 | 2 × 10−161 | 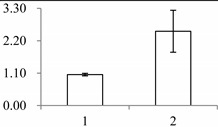 |
| I | 412 | Oxygen-evolving enhancer protein 1-2, chloroplastic | Oryza sativa subsp. japonica | Q9S841 | 131 | 485 | 3 × 10−169 | 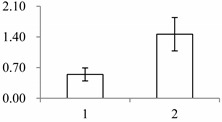 |
| I | 554 | Guanine nucleotide-binding protein subunit beta-like protein A | Oryza sativa subsp. japonica | P49027 | 93 | 521 | 0 | 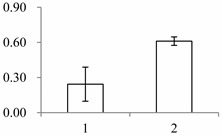 |
| II | 141 | SMALLER WITH VARIABLE BRANCHES (SVB) | Arabidopsis thaliana | Q9FXB0 | 274 | 164 | 6 × 10−48 | 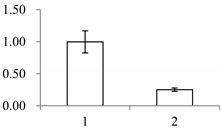 |
| II | 334 | NAD(P)-binding Rossmann-fold-containing protein | Arabidopsis thaliana | O80934 | 58 | 424 | 2 × 10−144 | 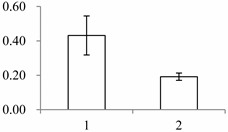 |
| II | 393 | Leucine-rich repeat extensin-like protein 3 | Arabidopsis thaliana | Q9T0K5 | 291 | 53.1 | 4 × 10−11 | 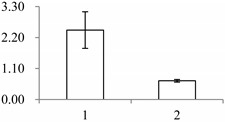 |
| II | 430 | Xyloglucan endotransglucosylase/hydrolase protein 22 | Arabidopsis thaliana | Q38857 | 262 | 420 | 1 × 10−144 | 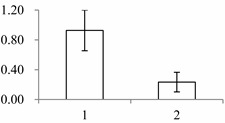 |
| II | 449 | Mannose-specific lectin | Allium sativum | P83886 | 239 | 63.2 | 3 × 10−11 | 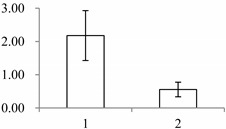 |
| II | 591 | NADP-dependent alkenal double bond reductase P2 | Arabidopsis thaliana | Q39173 | 65 | 479 | 2 × 10−166 | 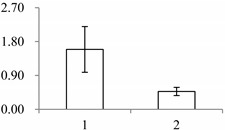 |
| II | 940 | NADP-dependent malic enzyme, chloroplastic (maeB) | Oryza sativa subsp. japonica | P43279 | 60 | 926 | 0 | 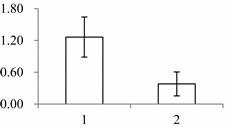 |
| II | 972 | Putative nucleic acid binding protein | Oryza sativa subsp. japonica | Q8S7G1 | 134 | 170 | 3 × 10−67 | 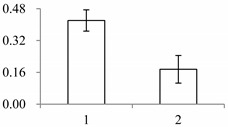 |
| II | 1028 | Putative elongation factor | Oryza sativa subsp. japonica | Q9ASR1 | 240 | 1524 | 0 | 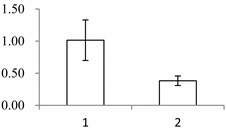 |
| II | 1090 | serine carboxypeptidase (SCPL)-like 18 | Brachypodium distachyon | XP_003560245.1 | 48 | 440 | 2 × 10−144 | 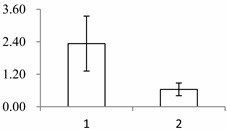 |
| II | 1146 | Proteasome subunit alpha type-7-B | Arabidopsis thaliana | O24616 | 181 | 443 | 3 × 10−156 | 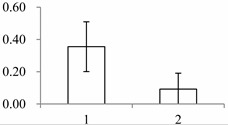 |
| II | 1201 | Quinone oxidoreductase-like protein At1g23740, chloroplastic | Arabidopsis thaliana | Q9ZUC1 | 104 | 489 | 1 × 10−166 | 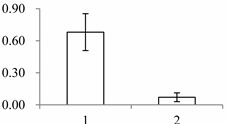 |
| III | 1535 | Cytosolic ascorbate peroxidase 1 | Gossypium mexicanum | A7KIX5 | 242 | 439 | 4 × 10−152 | 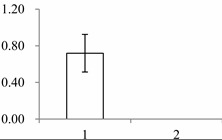 |
| III | 1545 | Xyloglucan endotransglucosylase/hydrolase, putative, expressed (XET/XTH) | Oryza sativa subsp. japonica | Q2R336 | 415 | 486 | 3 × 10−170 | 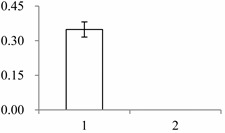 |
| III | 1547 | Beta-glucosidase 12 | Oryza sativa subsp. japonica | Q7XKV4 | 112 | 562 | 0 | 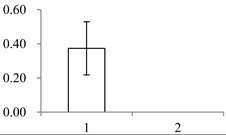 |
a Group I has the proteins down-regulated in labellum, group II has the protein up-regulated in labellum, and group III has the protein specifically expressed in the labellum; b Numbering corresponds to the 2-DE gel in Figure 2; c Accession numbers from the UniProt database; d The X-axis represents two parts of floral organ, and 1 and 2 represent labellum and inner lateral petal, respectively, and the Y-axis denotes the relative protein expression levels (normalized volume of spots), and the values are expressed as the mean of three replicates ± the standard deviation.
