Figure 2.
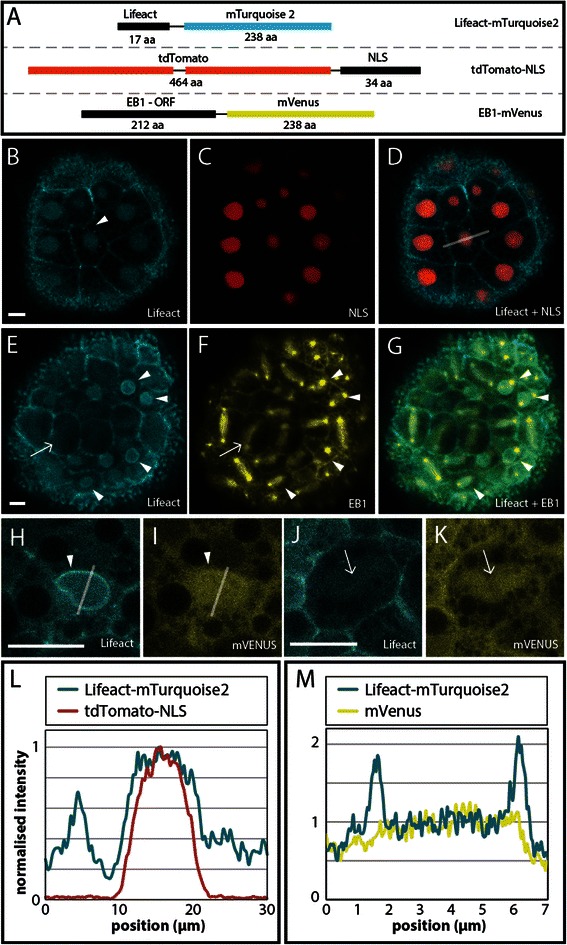
Localization of Lifeact-mTurquoise2, tdTomato-NLS and EB1-mVenus in early cleavage embryos. A) Messenger RNA for three different fluorescent probes was injected in pairs into embryos of Nematostella vectensis. B-D) In the same embryo Lifeact-mTurquoise2 is localized to cell boundaries and overlaps with tdTomato-NLS around the nucleus. E-G) In dividing cells EB1-mVenus is clearly localized at mitotic spindle fibers and centrosomes. Interestingly Lifeact-mTurquoise2 (E) exhibits a brighter fluorescent ring around the nucleus in a subset of cells (white arrowhead) that start to form centrosomes adjacent to the nucleus (G). Lifeact-mTurquoise2 is also visible as striations corresponding to the location of spindle fibers labeled with EB1-mVenus (white arrows in E and F). H-I) Double expression of Lifeact-mTurquoise2 and mVenus in Nematostella embryos shows high concentration of Lifeact-mTurquoise2 around the nucleus (H) compared to mVenus used as a control (I) (white arrowheads). J-K) Both Lifeact-mTurquoise2 and mVenus appear as striations with the same morphology as spindle fibers (white arrows). L) Quantification of fluorescent i3ntensity of Lifeact-mTurquoise2 and tdTomato-NLS in nuclei of early cleavage stage embryos (the x-axis is represented as a white line in D). M) Quantification of fluorescent intensity of Lifeact-mTurquoise2 and untagged mVenus in nuclei of early cleavage stage embryos (the x-axis is represented as a white line in H and I). See main text for a more detailed description. (Scale bar =10 μm in each image)
