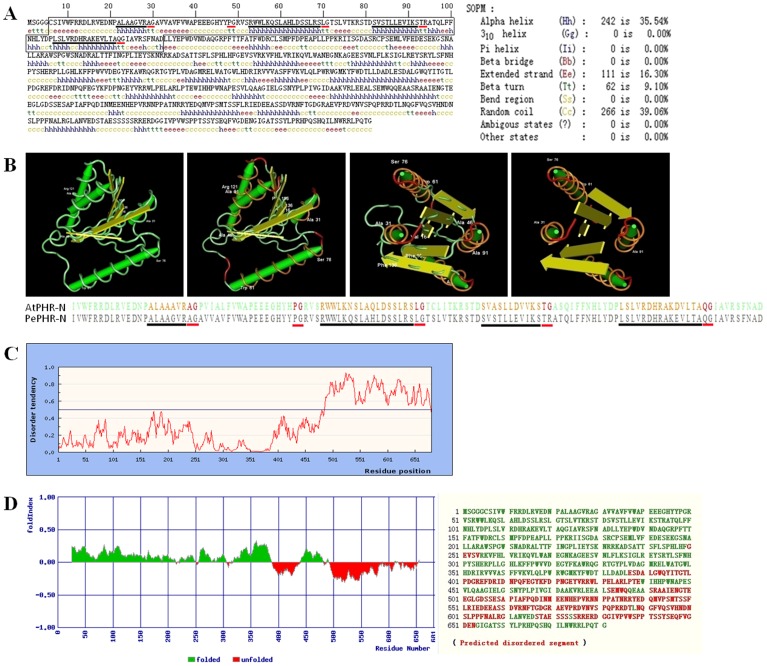
Figure 2. Structural analysis of the PeCRY1 protein.
(A) The secondary structure of PeCRY1 solved by the self-optimized prediction method (SPOM). The black box indicates the PeCRY1 PHR-N domain corresponding to that of AtCRY1. Black and red lines beneath the sequence indicate the α-helices and the β-turns in the PHR-N domain. (B) Comparison of the predicted three-dimensional structures of PeCRY1 and AtCRY1 using Cn3D software. Orange and red segments in the images correspond to the sequences marked by black and red lines, respectively, in (A). (C) Local disorder tendency of the PeCRY1 CCE domain based on an estimated-amino-acid-pairwise-energy-content analysis using IUPred software. (D) The fold disordering character of PeCRY1 predicted using FoldIndex software.